Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121837 |
| Name | oriT_pRetIE4771b |
| Organism | Rhizobium etli bv. mimosae str. IE4771 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP006988 (370942..370983 [+], 42 nt) |
| oriT length | 42 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 42 nt
>oriT_pRetIE4771b
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACGCCCT
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACGCCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file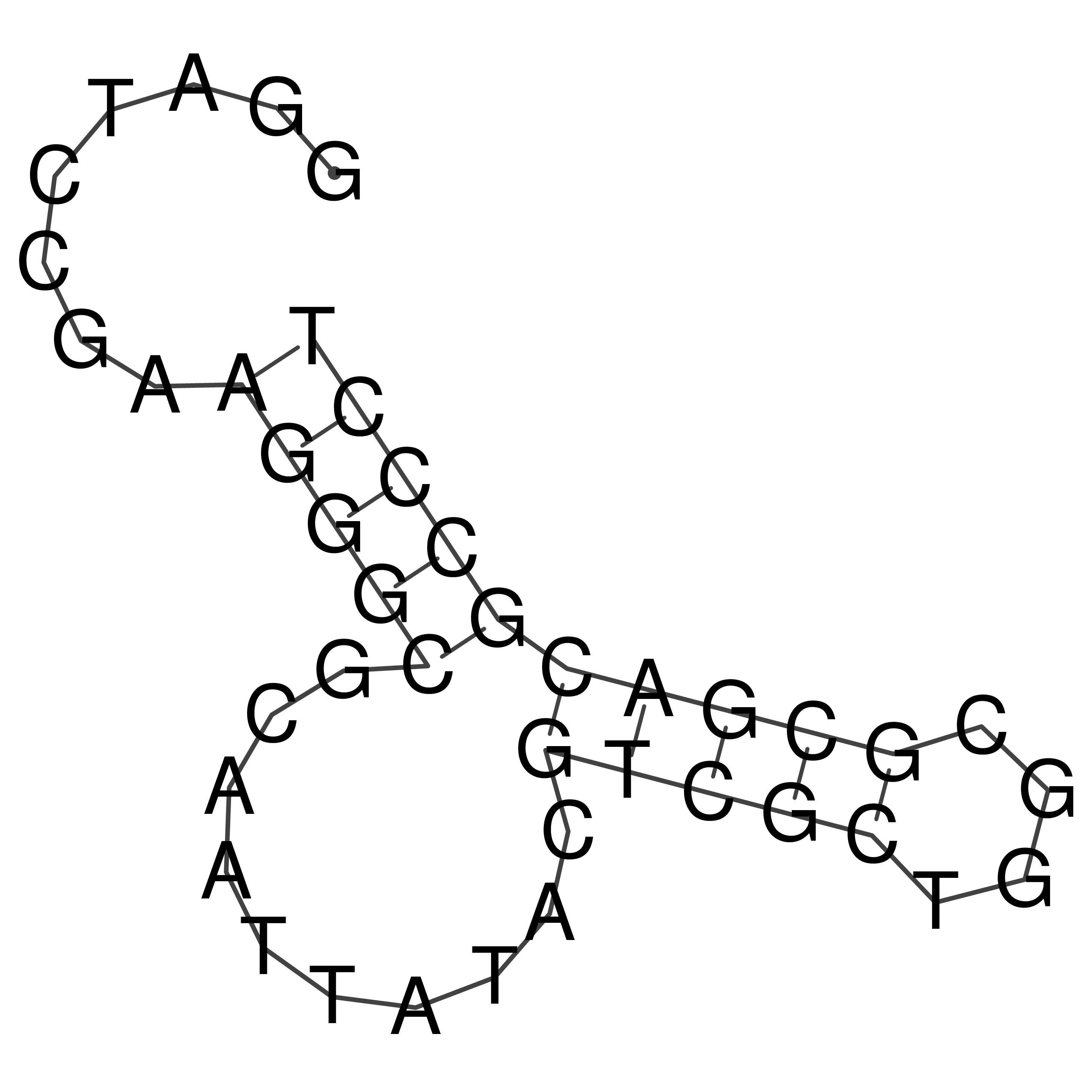
Relaxase
| ID | 13782 | GenBank | WP_040140651 |
| Name | traA_IE4771_RS24220_pRetIE4771b |
UniProt ID | A0A060I4Y1 |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122414.44 Da Isoelectric Point: 9.5561
>WP_040140651.1 Ti-type conjugative transfer relaxase TraA [Rhizobium etli]
MAVPHFSVSVVARGSGRSAVLSAAYRHCTKMEFEREARTIDYTRKQGLLHEEFVVPTDAPEWLRSMIADR
SVSGASEAFWNRVEDFEKRSDAQLAKDVTIALPIELTAEQNIALVRDFVEQHITVKGMVADWVYHDAPGN
PHVHLMTTLRPLTADGFGAKKIAVTGPDGNPIRNDAGKIVYELWAGSIDDFNTFRDGWFACQNRHLALAG
LDIYVDGRSFEKQGIELEPTIHLGVGTKAIERKSDGASDPVELERLELQEARRSENVRRIDRNPELVLAL
IMREKSVFDERDVAKILHRYVDDAALFQSLMVRILLCPETFRIECERIDLATGIRVPAKYTTRDMIRLEA
EMANRAVWLSQRSSHGVRDAVLAATFERHERLSDEQRTAIEHVAGPERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGAALAGKAAEGLEKEAGIISRTLASWELRWNQGRDQLDGKTVFVLDEAGMVSSRQMALFVE
AVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGKVGRAVDAYR
ANGKIIGSDLKADAVDNLIAAWDRDYDPAKTSLILAHLRRDVRMLNQMARIKLIERGVLDQGTMFKTADG
GRNFAVGDQIVFLKNEGSLGVKNGMLAKVVETGPGRITARIGERENARQVLVEQRFYNNLDHGYATTIHK
SQGATIDQVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKNGGLIPMLSRRNAKETTLDYEKSDFY
RQALRFAEARGLHLVNVARTLVRDRLEWVVSQKQKLANLGARLAAVAGKLGLIRGAAQSPTQNNTKEAKP
MVSGITTFAKSVDQAIEDKIAADPGLKKQWEEVSTRFHLVYAQPESAFKAVNVDAMLKDETAQKSTLAKL
GSEPESFGALKGKTGILASRSDRQEREKAGTNAPALARNLERYLRQRAEAERKFEVEERAVRLKVSIDIP
ALSPNAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLSLAAKEPNGQTFSA
VTSGMTAGQKTEVQSAWNSMRTVQQLAAHERTTEALKQAETLRQAKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCTKMEFEREARTIDYTRKQGLLHEEFVVPTDAPEWLRSMIADR
SVSGASEAFWNRVEDFEKRSDAQLAKDVTIALPIELTAEQNIALVRDFVEQHITVKGMVADWVYHDAPGN
PHVHLMTTLRPLTADGFGAKKIAVTGPDGNPIRNDAGKIVYELWAGSIDDFNTFRDGWFACQNRHLALAG
LDIYVDGRSFEKQGIELEPTIHLGVGTKAIERKSDGASDPVELERLELQEARRSENVRRIDRNPELVLAL
IMREKSVFDERDVAKILHRYVDDAALFQSLMVRILLCPETFRIECERIDLATGIRVPAKYTTRDMIRLEA
EMANRAVWLSQRSSHGVRDAVLAATFERHERLSDEQRTAIEHVAGPERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGAALAGKAAEGLEKEAGIISRTLASWELRWNQGRDQLDGKTVFVLDEAGMVSSRQMALFVE
AVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGKVGRAVDAYR
ANGKIIGSDLKADAVDNLIAAWDRDYDPAKTSLILAHLRRDVRMLNQMARIKLIERGVLDQGTMFKTADG
GRNFAVGDQIVFLKNEGSLGVKNGMLAKVVETGPGRITARIGERENARQVLVEQRFYNNLDHGYATTIHK
SQGATIDQVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKNGGLIPMLSRRNAKETTLDYEKSDFY
RQALRFAEARGLHLVNVARTLVRDRLEWVVSQKQKLANLGARLAAVAGKLGLIRGAAQSPTQNNTKEAKP
MVSGITTFAKSVDQAIEDKIAADPGLKKQWEEVSTRFHLVYAQPESAFKAVNVDAMLKDETAQKSTLAKL
GSEPESFGALKGKTGILASRSDRQEREKAGTNAPALARNLERYLRQRAEAERKFEVEERAVRLKVSIDIP
ALSPNAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLSLAAKEPNGQTFSA
VTSGMTAGQKTEVQSAWNSMRTVQQLAAHERTTEALKQAETLRQAKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A060I4Y1 |
T4CP
| ID | 16114 | GenBank | WP_040140646 |
| Name | traG_IE4771_RS24205_pRetIE4771b |
UniProt ID | _ |
| Length | 658 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 658 a.a. Molecular weight: 70940.14 Da Isoelectric Point: 9.5637
>WP_040140646.1 Ti-type conjugative transfer system protein TraG [Rhizobium etli]
MNRIVLFVAPVALMTVAAVCLTGIEHWLSAFGKTEAAHQTLGRVGIAFPYLAAALVGIVFLFASAGAIRI
KAAGWGVIAGGVATILIAAIRETIRLSALADKVPAGKSILSYVDPATLIGAGAAAMASCFALRVALAGNA
PFTSAAPKRIRGKRALHGEANWMKLPEAATLFSDTGGIVVGERYRVDKDSTAARAFRADERETWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLIVLDPSNEVAPMVSAHRTSANRDLFVLDPK
SPETGFNALDWIGKFGGTKEEDIASVASWIMSDSGGTRGVRDDFFRASALQLLTAMIADVCLSGHTEKVK
QTLRQVRANLSEPEPKLRQRLQEIYDNSDSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYQNYAAL
VSGKTFSTGDLAVGSTDVFINIDLRTLETHAGLARVIIGSFLNAIYNRDGSMKGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGHDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIIFTAGNSPLRCGRAIWFRRDDMKACVGTN
RFHQLKGGPEANLIEPARSAASKADREK
MNRIVLFVAPVALMTVAAVCLTGIEHWLSAFGKTEAAHQTLGRVGIAFPYLAAALVGIVFLFASAGAIRI
KAAGWGVIAGGVATILIAAIRETIRLSALADKVPAGKSILSYVDPATLIGAGAAAMASCFALRVALAGNA
PFTSAAPKRIRGKRALHGEANWMKLPEAATLFSDTGGIVVGERYRVDKDSTAARAFRADERETWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLIVLDPSNEVAPMVSAHRTSANRDLFVLDPK
SPETGFNALDWIGKFGGTKEEDIASVASWIMSDSGGTRGVRDDFFRASALQLLTAMIADVCLSGHTEKVK
QTLRQVRANLSEPEPKLRQRLQEIYDNSDSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYQNYAAL
VSGKTFSTGDLAVGSTDVFINIDLRTLETHAGLARVIIGSFLNAIYNRDGSMKGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGHDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIIFTAGNSPLRCGRAIWFRRDDMKACVGTN
RFHQLKGGPEANLIEPARSAASKADREK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16115 | GenBank | WP_040140668 |
| Name | t4cp2_IE4771_RS24300_pRetIE4771b |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91762.62 Da Isoelectric Point: 6.2213
>WP_040140668.1 conjugal transfer protein TrbE [Rhizobium etli]
MVALTRFRAAGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATVLERNELSRQINTILSRLG
SGWMIQVEAVRIPTFDYPSDDRCHFPDAVTRAIDAERRAHFGREQGHFESKHALILTYRPLESKKTALSK
YIYSDDESRKKTYADTVLFIFKNAVREIEQYFANTLSIRRMETRETFERGGERVARYDELLQFVRFCVTG
ENHPIRLPDAPMYLDWLATAELEHGLTPKVESRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFLF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVIVL
FDHDREALQEKAEAIRRLIQAEGFGARIETLNATEAFLGSLPGNWYCNIREPLINTSNLADLVPLNSVWS
GNPTAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGSALLPLTLAVGGEHYEIGGDHNGAERALAFCPLSELSNDADRAWATEWIEMLLALQGVAITPDHRN
AISRQIGLMASAAGRSLSDFVSGVQMRVIKDALHHYTVDGPMGQLLDAEQDGLSPVAVQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRAKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYQRIGFNERQIEIVSNAIPKREYYVVTPDGRRLFDMALGP
VALSFVGASGKEDLKRIRALRSEHGRDWPIHWLQSRGVHDAASLLNFE
MVALTRFRAAGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATVLERNELSRQINTILSRLG
SGWMIQVEAVRIPTFDYPSDDRCHFPDAVTRAIDAERRAHFGREQGHFESKHALILTYRPLESKKTALSK
YIYSDDESRKKTYADTVLFIFKNAVREIEQYFANTLSIRRMETRETFERGGERVARYDELLQFVRFCVTG
ENHPIRLPDAPMYLDWLATAELEHGLTPKVESRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFLF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVIVL
FDHDREALQEKAEAIRRLIQAEGFGARIETLNATEAFLGSLPGNWYCNIREPLINTSNLADLVPLNSVWS
GNPTAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGSALLPLTLAVGGEHYEIGGDHNGAERALAFCPLSELSNDADRAWATEWIEMLLALQGVAITPDHRN
AISRQIGLMASAAGRSLSDFVSGVQMRVIKDALHHYTVDGPMGQLLDAEQDGLSPVAVQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRAKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYQRIGFNERQIEIVSNAIPKREYYVVTPDGRRLFDMALGP
VALSFVGASGKEDLKRIRALRSEHGRDWPIHWLQSRGVHDAASLLNFE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 376108..389218
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IE4771_RS24225 (IE4771_PB00378) | 374371..374937 | + | 567 | WP_080711056 | conjugative transfer signal peptidase TraF | - |
| IE4771_RS24230 (IE4771_PB00379) | 374927..376090 | + | 1164 | WP_040140653 | conjugal transfer protein TraB | - |
| IE4771_RS24235 (IE4771_PB00380) | 376108..376719 | + | 612 | WP_040140655 | TraH family protein | virB1 |
| IE4771_RS24240 (IE4771_PB00381) | 376734..377111 | - | 378 | WP_040140657 | DUF5615 family PIN-like protein | - |
| IE4771_RS24245 (IE4771_PB00382) | 377108..377740 | - | 633 | WP_040140659 | DUF433 domain-containing protein | - |
| IE4771_RS24250 (IE4771_PB00383) | 377950..378171 | - | 222 | WP_010046230 | helix-turn-helix domain-containing protein | - |
| IE4771_RS24255 (IE4771_PB00384) | 378368..378685 | + | 318 | WP_010046233 | transcriptional repressor TraM | - |
| IE4771_RS24260 (IE4771_PB00385) | 378689..379393 | - | 705 | WP_040140791 | autoinducer binding domain-containing protein | - |
| IE4771_RS24265 (IE4771_PB00386) | 379715..381016 | - | 1302 | WP_040140660 | IncP-type conjugal transfer protein TrbI | virB10 |
| IE4771_RS24270 (IE4771_PB00387) | 381028..381465 | - | 438 | WP_010060774 | conjugal transfer protein TrbH | - |
| IE4771_RS24275 (IE4771_PB00388) | 381469..382281 | - | 813 | WP_040140662 | P-type conjugative transfer protein TrbG | virB9 |
| IE4771_RS24280 (IE4771_PB00389) | 382298..382960 | - | 663 | WP_010060772 | conjugal transfer protein TrbF | virB8 |
| IE4771_RS24285 (IE4771_PB00390) | 382985..384163 | - | 1179 | WP_040140666 | P-type conjugative transfer protein TrbL | virB6 |
| IE4771_RS24290 (IE4771_PB00391) | 384166..384342 | - | 177 | WP_010060768 | entry exclusion protein TrbK | - |
| IE4771_RS24295 (IE4771_PB00392) | 384336..385139 | - | 804 | WP_040140667 | P-type conjugative transfer protein TrbJ | virB5 |
| IE4771_RS24300 (IE4771_PB00393) | 385111..387567 | - | 2457 | WP_040140668 | conjugal transfer protein TrbE | virb4 |
| IE4771_RS24305 (IE4771_PB00394) | 387576..387875 | - | 300 | WP_040140669 | conjugal transfer protein TrbD | virB3 |
| IE4771_RS24310 (IE4771_PB00395) | 387868..388251 | - | 384 | WP_040140671 | TrbC/VirB2 family protein | virB2 |
| IE4771_RS24315 (IE4771_PB00396) | 388241..389218 | - | 978 | WP_040140672 | P-type conjugative transfer ATPase TrbB | virB11 |
| IE4771_RS24320 (IE4771_PB00397) | 389238..389855 | - | 618 | WP_040140673 | acyl-homoserine-lactone synthase TraI | - |
| IE4771_RS24325 (IE4771_PB00399) | 390243..391457 | + | 1215 | WP_040140795 | plasmid partitioning protein RepA | - |
| IE4771_RS24330 (IE4771_PB00400) | 391454..392479 | + | 1026 | WP_040140674 | plasmid partitioning protein RepB | - |
| IE4771_RS24335 (IE4771_PB00401) | 392635..393846 | + | 1212 | WP_010058593 | plasmid replication protein RepC | - |
Host bacterium
| ID | 22264 | GenBank | NZ_CP006988 |
| Plasmid name | pRetIE4771b | Incompatibility group | - |
| Plasmid size | 393846 bp | Coordinate of oriT [Strand] | 370942..370983 [+] |
| Host baterium | Rhizobium etli bv. mimosae str. IE4771 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | LinG, dxnH, linJ |
| Symbiosis gene | nifA, nifH, mLTONO_5203, nodD, fixN, fixO, nodA, nodB, nodC, nodS, nodU, nodI, nodJ, nodH, nifT, nifB, fixX, fixC, fixB, fixA, nifW, nifS, nifD, nifK, nifE, nifX |
| Anti-CRISPR | AcrIC6 |