Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121812 |
| Name | oriT_p2562_2 |
| Organism | Klebsiella michiganensis strain 2563 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP116216 (155113..155162 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_p2562_2
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file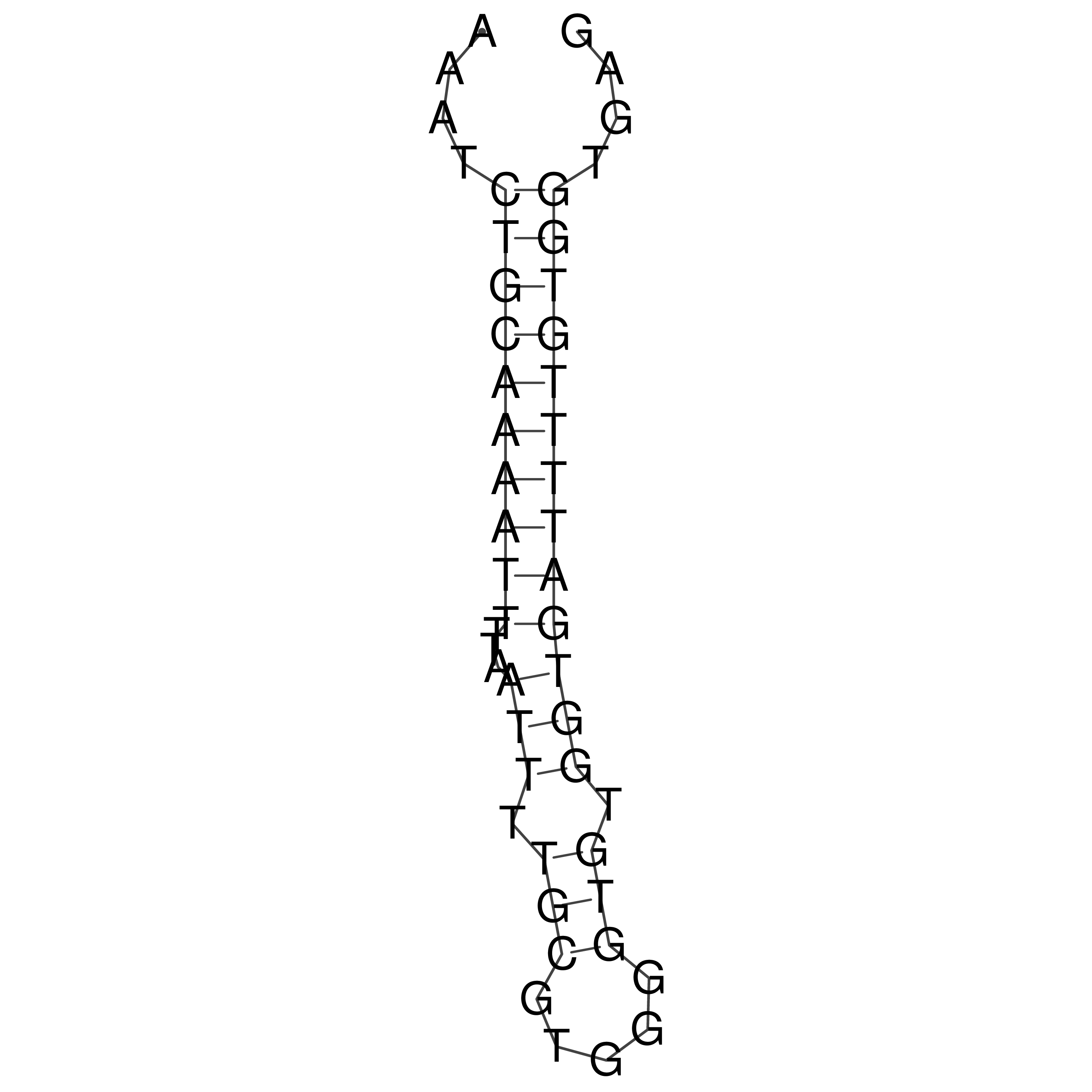
Relaxase
| ID | 13769 | GenBank | WP_040118408 |
| Name | traI_PHA56_RS30625_p2562_2 |
UniProt ID | _ |
| Length | 1754 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1754 a.a. Molecular weight: 190278.01 Da Isoelectric Point: 5.9989
>WP_040118408.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella/Raoultella group]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKPVDKAVFTEILKGKLPDGSDLTRM
QDGTNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTKALQQVETLAATRVMTDGKSETVLTGNL
IVAKFTHDTNRNEEPQLHTHAVVMNATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKIYREELKPLVE
ILGYKTEVVGKHGMWEMNEVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKAAIDPAEKMVEWMN
TMKETGFDMRSYREDADKRAAELASAPVSPVKTDGPDISDVVTKAIAGLSDRKVQFTYADLLARTVSQLE
AKPGVFEQARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVSVTPDASVVRQAP
FSDAVSVLAQDRPAMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQVTADIISEPDKGARYSLLAQEFAASVREGQESVAQISGTREQNVLNSVIRDTLKNEGVLGEKE
MTVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLKDKEGECLDMKVSAV
DSQWTLFRADKLPVAEGERLAVLGKIPDTRLKGGESITVLKSGDGQLTVQRPGQKTTQSLSVGSSPFDGI
EVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTMEKLARHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLSVPLLESDDLTFTRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVSVSSGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTGLTAGQRASTR
MILETPDRFTVVQGYAGVGKTTQFRAVTSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTLLLLLRNGQTPDFSNTLFLIDESSMVGLSDKAKALSLIAAGGGRAVSSGDTDQLQSISPGQPFRLMQQ
RSAADVAIMKEIVRQVPELRPAVYSLIDRDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTRAQEKAI
QEALKKGESVPAGQPATLYEALVKDYAGRTPEAQSQTLVITHLNKDRRALNGLIHDARRENGETGKEEIT
LPVLVTSNIRDGELRRLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVT
LYRREDITVSEGDRMRFSKSDPERGYVANSVWEVKSVSGDSVTFSDGKTSRTLNPKADEAQRHIDLAYAI
TAHGAQGASEPFAIALEGVAEGRKAMASFESAYVGLSRMKQHVQVYTDSREGWIKAINASPSKATAHDIL
APRNDRAVHTAEQLFSRARPMNETAAGRAALQQSGLAKGISPGKFISPGKQYPQPHVALPVYDKNGKEAG
IWLSSLTDSDGRLQVMGGVGRVMGNEDARFVALQGSRNGESLLAGNMGEGVRVARDNPDSGVVVRLAGDD
RPWNPGAITGGRVWADPIPVVSPEAAGTDIPLPPEVLAQRAAEEAQRQNMEKQAEQTAREVAGDGRKAGE
SEDRVKEVIGDVIRGLEREKPGEEKMTLPDDPQTRKQEAAVQQVAQESLQRERLQQMERDMVRDLNREKT
LGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKPVDKAVFTEILKGKLPDGSDLTRM
QDGTNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTKALQQVETLAATRVMTDGKSETVLTGNL
IVAKFTHDTNRNEEPQLHTHAVVMNATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKIYREELKPLVE
ILGYKTEVVGKHGMWEMNEVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKAAIDPAEKMVEWMN
TMKETGFDMRSYREDADKRAAELASAPVSPVKTDGPDISDVVTKAIAGLSDRKVQFTYADLLARTVSQLE
AKPGVFEQARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVSVTPDASVVRQAP
FSDAVSVLAQDRPAMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQVTADIISEPDKGARYSLLAQEFAASVREGQESVAQISGTREQNVLNSVIRDTLKNEGVLGEKE
MTVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLKDKEGECLDMKVSAV
DSQWTLFRADKLPVAEGERLAVLGKIPDTRLKGGESITVLKSGDGQLTVQRPGQKTTQSLSVGSSPFDGI
EVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTMEKLARHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLSVPLLESDDLTFTRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVSVSSGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTGLTAGQRASTR
MILETPDRFTVVQGYAGVGKTTQFRAVTSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTLLLLLRNGQTPDFSNTLFLIDESSMVGLSDKAKALSLIAAGGGRAVSSGDTDQLQSISPGQPFRLMQQ
RSAADVAIMKEIVRQVPELRPAVYSLIDRDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTRAQEKAI
QEALKKGESVPAGQPATLYEALVKDYAGRTPEAQSQTLVITHLNKDRRALNGLIHDARRENGETGKEEIT
LPVLVTSNIRDGELRRLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVT
LYRREDITVSEGDRMRFSKSDPERGYVANSVWEVKSVSGDSVTFSDGKTSRTLNPKADEAQRHIDLAYAI
TAHGAQGASEPFAIALEGVAEGRKAMASFESAYVGLSRMKQHVQVYTDSREGWIKAINASPSKATAHDIL
APRNDRAVHTAEQLFSRARPMNETAAGRAALQQSGLAKGISPGKFISPGKQYPQPHVALPVYDKNGKEAG
IWLSSLTDSDGRLQVMGGVGRVMGNEDARFVALQGSRNGESLLAGNMGEGVRVARDNPDSGVVVRLAGDD
RPWNPGAITGGRVWADPIPVVSPEAAGTDIPLPPEVLAQRAAEEAQRQNMEKQAEQTAREVAGDGRKAGE
SEDRVKEVIGDVIRGLEREKPGEEKMTLPDDPQTRKQEAAVQQVAQESLQRERLQQMERDMVRDLNREKT
LGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7603 | GenBank | WP_032700831 |
| Name | WP_032700831_p2562_2 |
UniProt ID | A0A485D480 |
| Length | 129 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 129 a.a. Molecular weight: 14705.63 Da Isoelectric Point: 4.4711
>WP_032700831.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Klebsiella/Raoultella group]
MARVQAYASDEVAEKINAIVEKRKAEGAKDKDVSFSSVASMLLELGLRVYEAQMERKESGFNQAEFNKML
LENVIKTQFSISKVLAMESLSPHIQGDERFDFKSMVMNIRDDAKEVVERFFPDTDDEES
MARVQAYASDEVAEKINAIVEKRKAEGAKDKDVSFSSVASMLLELGLRVYEAQMERKESGFNQAEFNKML
LENVIKTQFSISKVLAMESLSPHIQGDERFDFKSMVMNIRDDAKEVVERFFPDTDDEES
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A485D480 |
T4CP
| ID | 16094 | GenBank | WP_040118437 |
| Name | traD_PHA56_RS30630_p2562_2 |
UniProt ID | _ |
| Length | 776 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 776 a.a. Molecular weight: 86788.85 Da Isoelectric Point: 4.9734
>WP_040118437.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella/Raoultella group]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGRSLEYNAEQILADKYTIWCGEQLWTSFVLAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFATGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRSLDTRVDTRLSALLEAREAEGSLARALFTPDAPAPDPAEKDVKVGNTPEQSQPAE
SIESPATVKVPATAKTPAADEIGQSPETPEPAAPVTKVVTVPLIRPRSATATGTAAVATTAASTTSATTV
PAGGTEQELAQQPTEQGQDRLPAGMNDDGVIEDMDAYDAWLAGEQTQRDMQRREEVNINHSHRRDEQDDI
EIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGRSLEYNAEQILADKYTIWCGEQLWTSFVLAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFATGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRSLDTRVDTRLSALLEAREAEGSLARALFTPDAPAPDPAEKDVKVGNTPEQSQPAE
SIESPATVKVPATAKTPAADEIGQSPETPEPAAPVTKVVTVPLIRPRSATATGTAAVATTAASTTSATTV
PAGGTEQELAQQPTEQGQDRLPAGMNDDGVIEDMDAYDAWLAGEQTQRDMQRREEVNINHSHRRDEQDDI
EIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 126645..155715
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PHA56_RS30630 (PHA56_30630) | 126645..128975 | - | 2331 | WP_040118437 | type IV conjugative transfer system coupling protein TraD | virb4 |
| PHA56_RS30635 (PHA56_30635) | 129169..129900 | - | 732 | WP_032716670 | conjugal transfer complement resistance protein TraT | - |
| PHA56_RS30640 (PHA56_30640) | 130095..130709 | - | 615 | WP_032732092 | hypothetical protein | - |
| PHA56_RS30645 (PHA56_30645) | 130735..133557 | - | 2823 | WP_040118409 | conjugal transfer mating-pair stabilization protein TraG | traG |
| PHA56_RS30650 (PHA56_30650) | 133557..134927 | - | 1371 | WP_032700807 | conjugal transfer pilus assembly protein TraH | traH |
| PHA56_RS30655 (PHA56_30655) | 134914..135342 | - | 429 | WP_032700808 | conjugal transfer protein TrbF | - |
| PHA56_RS30660 (PHA56_30660) | 135335..135901 | - | 567 | WP_032732090 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| PHA56_RS30665 (PHA56_30665) | 135876..136112 | - | 237 | WP_032700810 | type-F conjugative transfer system pilin chaperone TraQ | - |
| PHA56_RS30670 (PHA56_30670) | 136123..136875 | - | 753 | WP_040118410 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| PHA56_RS30675 (PHA56_30675) | 136898..137224 | - | 327 | WP_032700812 | hypothetical protein | - |
| PHA56_RS30680 (PHA56_30680) | 137514..138482 | - | 969 | WP_077255412 | IS5 family transposase | - |
| PHA56_RS30685 (PHA56_30685) | 138562..140517 | - | 1956 | WP_040118411 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| PHA56_RS30690 (PHA56_30690) | 140517..141146 | - | 630 | WP_032700817 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| PHA56_RS30695 (PHA56_30695) | 141159..142118 | - | 960 | WP_032716690 | conjugal transfer pilus assembly protein TraU | traU |
| PHA56_RS30700 (PHA56_30700) | 142159..142788 | - | 630 | WP_032716660 | type-F conjugative transfer system protein TraW | traW |
| PHA56_RS30705 (PHA56_30705) | 142785..143177 | - | 393 | WP_032716659 | type-F conjugative transfer system protein TrbI | - |
| PHA56_RS30710 (PHA56_30710) | 143177..145816 | - | 2640 | WP_032716658 | type IV secretion system protein TraC | virb4 |
| PHA56_RS30715 (PHA56_30715) | 145896..146285 | - | 390 | WP_040118412 | hypothetical protein | - |
| PHA56_RS30720 (PHA56_30720) | 146662..147066 | - | 405 | WP_040118414 | hypothetical protein | - |
| PHA56_RS30725 (PHA56_30725) | 147063..147284 | - | 222 | WP_032732073 | hypothetical protein | - |
| PHA56_RS30730 (PHA56_30730) | 147766..148245 | - | 480 | WP_223306708 | hypothetical protein | - |
| PHA56_RS30735 (PHA56_30735) | 148352..148921 | - | 570 | WP_071889124 | type IV conjugative transfer system lipoprotein TraV | traV |
| PHA56_RS30740 (PHA56_30740) | 148940..149146 | - | 207 | WP_032700826 | hypothetical protein | - |
| PHA56_RS30745 (PHA56_30745) | 149127..149723 | - | 597 | WP_119834738 | conjugal transfer protein TraP | - |
| PHA56_RS30750 (PHA56_30750) | 149716..151134 | - | 1419 | WP_040118415 | F-type conjugal transfer pilus assembly protein TraB | traB |
| PHA56_RS30755 (PHA56_30755) | 151134..151868 | - | 735 | WP_040118416 | type-F conjugative transfer system secretin TraK | traK |
| PHA56_RS30760 (PHA56_30760) | 151855..152421 | - | 567 | WP_040118417 | type IV conjugative transfer system protein TraE | traE |
| PHA56_RS30765 (PHA56_30765) | 152441..152746 | - | 306 | WP_020323523 | type IV conjugative transfer system protein TraL | traL |
| PHA56_RS30770 (PHA56_30770) | 152760..153128 | - | 369 | WP_047665734 | type IV conjugative transfer system pilin TraA | - |
| PHA56_RS30775 (PHA56_30775) | 153190..153354 | - | 165 | WP_032444506 | TraY domain-containing protein | - |
| PHA56_RS30780 (PHA56_30780) | 153532..154221 | - | 690 | WP_071889125 | hypothetical protein | - |
| PHA56_RS30785 (PHA56_30785) | 154422..154811 | - | 390 | WP_032700831 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| PHA56_RS30790 (PHA56_30790) | 155236..155715 | + | 480 | WP_032716648 | transglycosylase SLT domain-containing protein | virB1 |
| PHA56_RS30795 (PHA56_30795) | 155753..156283 | - | 531 | WP_032716647 | antirestriction protein | - |
| PHA56_RS30800 (PHA56_30800) | 156968..157114 | - | 147 | WP_032700834 | hypothetical protein | - |
| PHA56_RS30805 (PHA56_30805) | 157208..157555 | - | 348 | WP_032700835 | hypothetical protein | - |
| PHA56_RS30810 (PHA56_30810) | 157581..157739 | - | 159 | WP_162180130 | hypothetical protein | - |
| PHA56_RS30815 (PHA56_30815) | 157796..158071 | - | 276 | WP_032700867 | hypothetical protein | - |
| PHA56_RS30820 (PHA56_30820) | 158890..159171 | - | 282 | WP_004118961 | helix-turn-helix transcriptional regulator | - |
| PHA56_RS30825 (PHA56_30825) | 159152..159481 | - | 330 | WP_004118963 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| PHA56_RS30830 (PHA56_30830) | 159801..159888 | - | 88 | Protein_166 | theronine dehydrogenase | - |
| PHA56_RS30835 (PHA56_30835) | 159885..160613 | - | 729 | WP_004118966 | plasmid SOS inhibition protein A | - |
Host bacterium
| ID | 22239 | GenBank | NZ_CP116216 |
| Plasmid name | p2562_2 | Incompatibility group | IncFIB |
| Plasmid size | 183276 bp | Coordinate of oriT [Strand] | 155113..155162 [+] |
| Host baterium | Klebsiella michiganensis strain 2563 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE, arsR, arsD, arsA, arsB, arsC, arsH |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |