Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121741 |
| Name | oriT_pAt2 |
| Organism | Agrobacterium tumefaciens strain cl001 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP048577 (179086..179124 [+], 39 nt) |
| oriT length | 39 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 39 nt
>oriT_pAt2
GGATCCGAAGGGCGCAATTATACGTCGCTGATGCGACGC
GGATCCGAAGGGCGCAATTATACGTCGCTGATGCGACGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file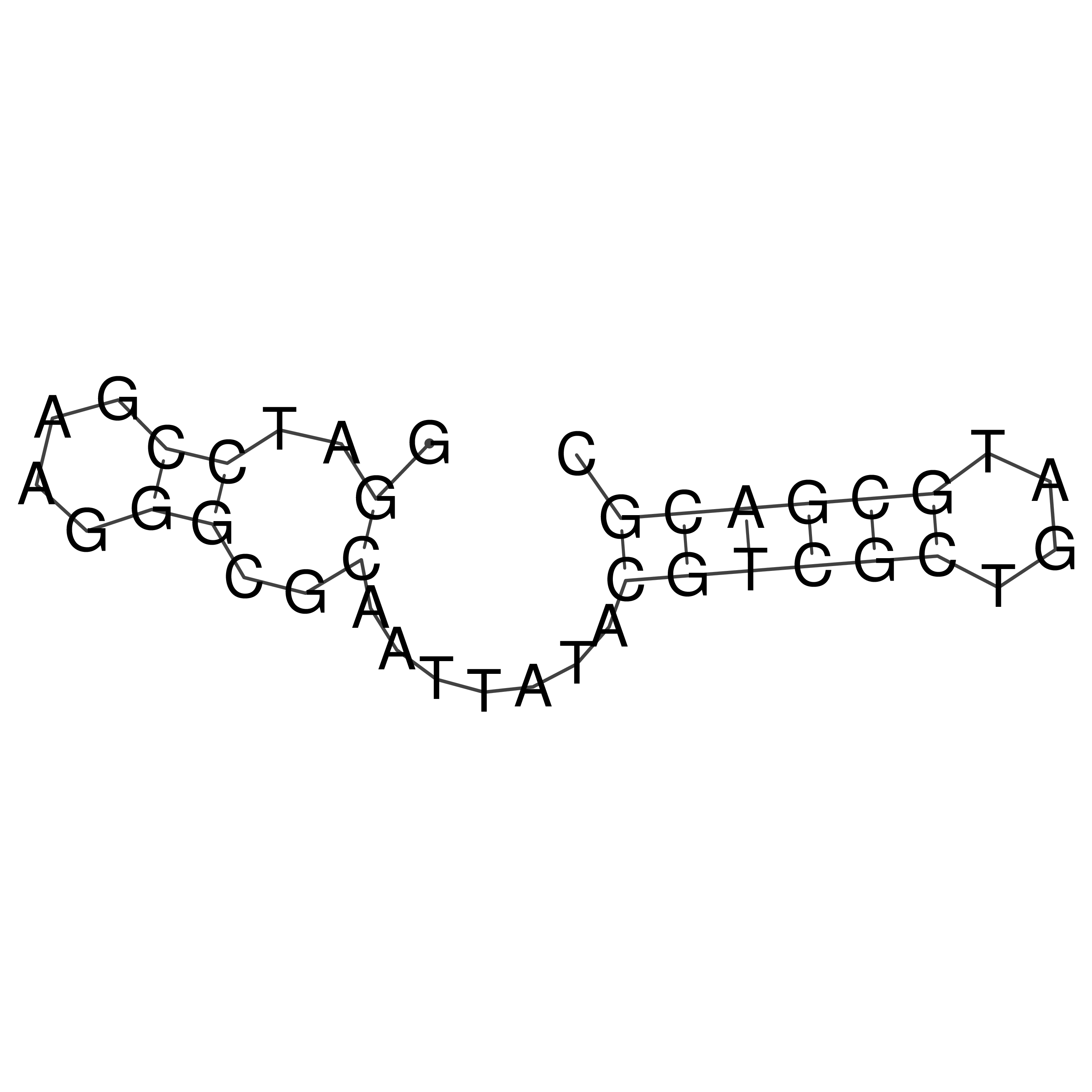
Relaxase
| ID | 13721 | GenBank | WP_333723780 |
| Name | traA_FY131_RS28000_pAt2 |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 122002.57 Da Isoelectric Point: 9.3614
>WP_333723780.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium tumefaciens]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKQGLLHEEFVIPADAPEWLQSLIADR
SVAGASEAFWNKVEDFEKRSDAQLAKDVTIALPIELSAEQNIELVRDFVARHITAHGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGSKKVVVTGPDGSVLRNDAGKIVYELWAGGLEDFNAFRDGWFSCQNRHLTLAG
LDIRVDGRSFEKQGIELTPTIHLGVGTKAIERKAESVEEKAALERLELQEERRAENARRIQRNPEIVIDL
ITREKSVFDERDVAKILHRYIDHAGLFQSLKARILQSPDALRLDRERLDFTTGVRAPAKYTTRELIRLEA
EMANRAIWLLQRSSHGVRQPVLEATFARHERLSGEQKAAIEHVTGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYHVVGGALAGKAAEGLEKEAGIVSRTLSSWELHWDQGRGQLDEKTVFVLDEAGMVSSRQMALFVE
TVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGHLSKAITAYQ
SNGKMIGSELKADAVTNLIADWNREYAPTKSTLILAHLRRDVRMLNELARAKLIERGIIEQGSPFKTADG
NRSFSAGEQIVFLKNEGSLGVKNGMLGKVVEAASGRLVAVIGEGEHLRKVIVEQRFYNNVDHGYATTVHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLGVYYGHRSFAKAGGLVEILSRRNSKETTLDYARGSIY
HQALRFAEGRGLHIVNVARTVARDRLEWTLRQKQKLVDLAGRLAAVGARLGFATSTANTIPTTAKEDKPM
VAGITTFPKSIDQAVEDKLASDPALKKQWEEVSMRFHHVYAQPESAFKAVDVDAMLSDPATAAKTLSHIT
AEPEAFGSLKGKAGVFASRADKSDRDRALKNVTPLADSISDYLRQRGDAERRNHAEELAVRRQVALEIPA
LSSNAKSVLERVRDAIDRNDLPSGLEFALADKMVKAELEGFAKAVTERFGERTFLSFAAKDANGEAFQRV
TSGMNAAQKSEVQQAWNTMRTVQQLSAHERSVTALKQTEALRQTKFQGLTLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKQGLLHEEFVIPADAPEWLQSLIADR
SVAGASEAFWNKVEDFEKRSDAQLAKDVTIALPIELSAEQNIELVRDFVARHITAHGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGSKKVVVTGPDGSVLRNDAGKIVYELWAGGLEDFNAFRDGWFSCQNRHLTLAG
LDIRVDGRSFEKQGIELTPTIHLGVGTKAIERKAESVEEKAALERLELQEERRAENARRIQRNPEIVIDL
ITREKSVFDERDVAKILHRYIDHAGLFQSLKARILQSPDALRLDRERLDFTTGVRAPAKYTTRELIRLEA
EMANRAIWLLQRSSHGVRQPVLEATFARHERLSGEQKAAIEHVTGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYHVVGGALAGKAAEGLEKEAGIVSRTLSSWELHWDQGRGQLDEKTVFVLDEAGMVSSRQMALFVE
TVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGHLSKAITAYQ
SNGKMIGSELKADAVTNLIADWNREYAPTKSTLILAHLRRDVRMLNELARAKLIERGIIEQGSPFKTADG
NRSFSAGEQIVFLKNEGSLGVKNGMLGKVVEAASGRLVAVIGEGEHLRKVIVEQRFYNNVDHGYATTVHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLGVYYGHRSFAKAGGLVEILSRRNSKETTLDYARGSIY
HQALRFAEGRGLHIVNVARTVARDRLEWTLRQKQKLVDLAGRLAAVGARLGFATSTANTIPTTAKEDKPM
VAGITTFPKSIDQAVEDKLASDPALKKQWEEVSMRFHHVYAQPESAFKAVDVDAMLSDPATAAKTLSHIT
AEPEAFGSLKGKAGVFASRADKSDRDRALKNVTPLADSISDYLRQRGDAERRNHAEELAVRRQVALEIPA
LSSNAKSVLERVRDAIDRNDLPSGLEFALADKMVKAELEGFAKAVTERFGERTFLSFAAKDANGEAFQRV
TSGMNAAQKSEVQQAWNTMRTVQQLSAHERSVTALKQTEALRQTKFQGLTLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16035 | GenBank | WP_333723668 |
| Name | traG_FY131_RS27985_pAt2 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69249.15 Da Isoelectric Point: 9.4654
>WP_333723668.1 Ti-type conjugative transfer system protein TraG [Agrobacterium tumefaciens]
MKANKIVLVILPGAMMVATMFAMTGIETWLAGFGKSEAAKLAVGRVGVAAPYLAAAALGLILLFATAGSA
SIRAAGVGAAIGSAAVILTACLREGVRLAALSAQVPAGESALSYLDPSTMIGAAAALMSGCFALRVAMVG
NAAFARTAPKRIIGKRALHGEADWMNLAEAGRLLPDSGGIVVGERYRVDRDGVAGVSFRADSSETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLIVLDPSNEVAPMVSKHREEAGRRVRILD
PRKPATGFNALDWVGQYGGTKEEDIASVASWIMSDSGRASGVRDDFFRASGLQLLTAIIADVCLSGHTPK
EKQTLRQVRENLSEPEPKLRQRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYQNYA
ALVSGSTFRTEDIASRKTDVFINIDLKTLETHAGLARVVIGSFLNAIYNRDGAMEGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQLRENYGGRDASSKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSLQSRGSSRTRSKQLAARPLIQQHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRKDMTACVE
RGRFGRREAPADEQ
MKANKIVLVILPGAMMVATMFAMTGIETWLAGFGKSEAAKLAVGRVGVAAPYLAAAALGLILLFATAGSA
SIRAAGVGAAIGSAAVILTACLREGVRLAALSAQVPAGESALSYLDPSTMIGAAAALMSGCFALRVAMVG
NAAFARTAPKRIIGKRALHGEADWMNLAEAGRLLPDSGGIVVGERYRVDRDGVAGVSFRADSSETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLIVLDPSNEVAPMVSKHREEAGRRVRILD
PRKPATGFNALDWVGQYGGTKEEDIASVASWIMSDSGRASGVRDDFFRASGLQLLTAIIADVCLSGHTPK
EKQTLRQVRENLSEPEPKLRQRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYQNYA
ALVSGSTFRTEDIASRKTDVFINIDLKTLETHAGLARVVIGSFLNAIYNRDGAMEGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQLRENYGGRDASSKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSLQSRGSSRTRSKQLAARPLIQQHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRKDMTACVE
RGRFGRREAPADEQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16036 | GenBank | WP_333723684 |
| Name | t4cp2_FY131_RS28080_pAt2 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91757.62 Da Isoelectric Point: 5.9703
>WP_333723684.1 conjugal transfer protein TrbE [Agrobacterium tumefaciens]
MAALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
TGWMIQVEAIRVPTVDYPTPGQCHFPDPVTRAIDAERRTHFGREQGHFESKHAIILTYRPLESKKTALAK
YVYSDEASRKKSYADTVLFMFRNAIREIEQYLANTLSIARMQTREVRERGGERVARYDDLMQFVRFCITG
ESHPVRLPEVPMYLDWLVTAELEHGLTPKVESRFLAVVAIDGLPAESWPGILNNLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVIL
FDEEREGLREKAEAIRRLVQAEGFGARVETLNATDAFLGSLPGNWYANIREPLINTSNLADLVPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEFAQIFA
FDKGNSLLPLTLAAGGDHYEIGNDQNGEGRALAFCPLFDLSTDGDRAWATEWVEMLVALQGVTITPDHRN
AISRQIGLMATAPGKSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLRAFQCFEIEQLMNMG
ERNLVPVLTYLFHRIEKRLDGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVSGAIPKREYYVATPDGRRLFDMSLGP
VALSFVGASGKEDLKLIRALKAEHGQDWPTHWLQSRGVHDATSHFNVQ
MAALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
TGWMIQVEAIRVPTVDYPTPGQCHFPDPVTRAIDAERRTHFGREQGHFESKHAIILTYRPLESKKTALAK
YVYSDEASRKKSYADTVLFMFRNAIREIEQYLANTLSIARMQTREVRERGGERVARYDDLMQFVRFCITG
ESHPVRLPEVPMYLDWLVTAELEHGLTPKVESRFLAVVAIDGLPAESWPGILNNLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVIL
FDEEREGLREKAEAIRRLVQAEGFGARVETLNATDAFLGSLPGNWYANIREPLINTSNLADLVPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEFAQIFA
FDKGNSLLPLTLAAGGDHYEIGNDQNGEGRALAFCPLFDLSTDGDRAWATEWVEMLVALQGVTITPDHRN
AISRQIGLMATAPGKSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLRAFQCFEIEQLMNMG
ERNLVPVLTYLFHRIEKRLDGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVSGAIPKREYYVATPDGRRLFDMSLGP
VALSFVGASGKEDLKLIRALKAEHGQDWPTHWLQSRGVHDATSHFNVQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 184252..197130
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FY131_RS28005 (FY131_27795) | 182545..183075 | + | 531 | WP_333723671 | conjugative transfer signal peptidase TraF | - |
| FY131_RS28010 (FY131_27800) | 183065..184237 | + | 1173 | WP_333723672 | conjugal transfer protein TraB | - |
| FY131_RS28015 (FY131_27805) | 184252..184860 | + | 609 | WP_333723673 | TraH family protein | virB1 |
| FY131_RS28020 (FY131_27810) | 184909..185286 | - | 378 | WP_333723674 | DUF5615 family PIN-like protein | - |
| FY131_RS28025 (FY131_27815) | 185291..185910 | - | 620 | Protein_174 | DUF433 domain-containing protein | - |
| FY131_RS28030 (FY131_27820) | 186030..186248 | - | 219 | WP_333723675 | helix-turn-helix domain-containing protein | - |
| FY131_RS28035 (FY131_27825) | 186387..186707 | + | 321 | WP_333723676 | transcriptional repressor TraM | - |
| FY131_RS28040 (FY131_27830) | 186709..187410 | - | 702 | WP_333723677 | autoinducer binding domain-containing protein | - |
| FY131_RS28045 (FY131_27835) | 187632..188930 | - | 1299 | WP_333723678 | IncP-type conjugal transfer protein TrbI | virB10 |
| FY131_RS28050 (FY131_27840) | 188944..189381 | - | 438 | WP_333723679 | conjugal transfer protein TrbH | - |
| FY131_RS28055 (FY131_27845) | 189385..190196 | - | 812 | Protein_180 | P-type conjugative transfer protein TrbG | - |
| FY131_RS28060 (FY131_27850) | 190213..190875 | - | 663 | WP_333723680 | conjugal transfer protein TrbF | virB8 |
| FY131_RS28065 (FY131_27855) | 190897..192075 | - | 1179 | WP_333723681 | P-type conjugative transfer protein TrbL | virB6 |
| FY131_RS28070 (FY131_27860) | 192069..192272 | - | 204 | WP_333723682 | entry exclusion protein TrbK | - |
| FY131_RS28075 (FY131_27865) | 192269..193072 | - | 804 | WP_333723683 | P-type conjugative transfer protein TrbJ | virB5 |
| FY131_RS28080 (FY131_27870) | 193044..195500 | - | 2457 | WP_333723684 | conjugal transfer protein TrbE | virb4 |
| FY131_RS28085 (FY131_27875) | 195511..195795 | - | 285 | WP_333723685 | conjugal transfer protein TrbD | virB3 |
| FY131_RS28090 (FY131_27880) | 195792..196169 | - | 378 | WP_333723686 | TrbC/VirB2 family protein | virB2 |
| FY131_RS28095 (FY131_27885) | 196159..197130 | - | 972 | WP_333723687 | P-type conjugative transfer ATPase TrbB | virB11 |
| FY131_RS28100 (FY131_27890) | 197133..197759 | - | 627 | WP_333723688 | acyl-homoserine-lactone synthase TraI | - |
Host bacterium
| ID | 22168 | GenBank | NZ_CP048577 |
| Plasmid name | pAt2 | Incompatibility group | - |
| Plasmid size | 198129 bp | Coordinate of oriT [Strand] | 179086..179124 [+] |
| Host baterium | Agrobacterium tumefaciens strain cl001 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |