Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121740 |
| Name | oriT_pQ15_94_4 |
| Organism | Agrobacterium tumefaciens strain Q15/94 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP049221 (126850..126893 [-], 44 nt) |
| oriT length | 44 nt |
| IRs (inverted repeats) | 21..27, 32..38 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 44 nt
>oriT_pQ15_94_4
GGATCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
GGATCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file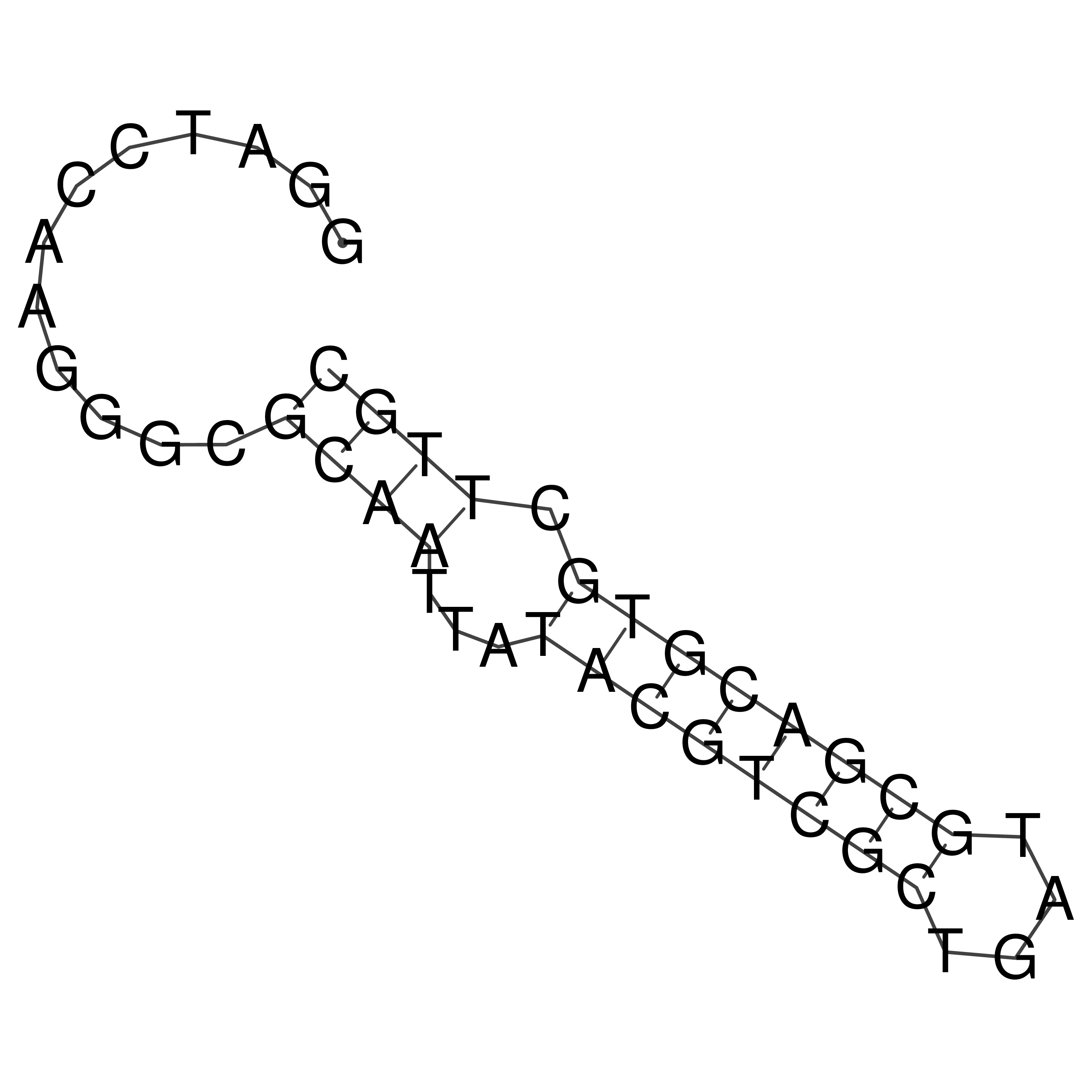
Relaxase
| ID | 13720 | GenBank | WP_333722975 |
| Name | traA_G6M86_RS29360_pQ15_94_4 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123806.33 Da Isoelectric Point: 9.7079
>WP_333722975.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium tumefaciens]
MAIAHFSASIVSRGGGRSVVLSAAYRHCAKMEYEREARTIDYTRKEGLVHEEFMLPADAPKWVRAMIADR
SVAGASEAFWNKVEAFEKRSDAQLARDLTIALPVELSSEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEDGFGSKKVAVIGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYEKQGIELEPTIHLGVGAKAIERKAQEQGVRPELERIELNEQRRSENTRRILRNPGIVVDL
ITREKSVFDERDVAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMVRQALWLSNRDGHAVFEAALDATFRRHERLSQEQKTAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDKAVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDQSKTTLILAHLRRDVRTLNIMAREKLVERGIVGEGHVFKTADG
IRQFDAGDQIVFLKNEGSLGVKNGMIAHVIEAAPNRIIAVVGEGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGRLY
REALRFAESRGLNIVQVARTLVRDRLDWTLRQKTKLVDLGQRLAVFAERLGLHHPPKTQTMKEAAPMVAG
IKTFSGSVADTVENKLDTDPALKKQWEEVSARFAYVFADPETAFRAMNFDAVLTEKDTARQVVQKLEAEP
ASIGPLRGKTGILASKTEREDRRIAEINVPALKRDLEQYLQMREAAIKRLSTDEQVQRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTQRFGERTLLTNAAREPSGKLFDELSDG
MQPEQKERLKEAWPIMRTSQQLTAHERTVQTLRQAEDLRLSQRQTPVMKQ
MAIAHFSASIVSRGGGRSVVLSAAYRHCAKMEYEREARTIDYTRKEGLVHEEFMLPADAPKWVRAMIADR
SVAGASEAFWNKVEAFEKRSDAQLARDLTIALPVELSSEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEDGFGSKKVAVIGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYEKQGIELEPTIHLGVGAKAIERKAQEQGVRPELERIELNEQRRSENTRRILRNPGIVVDL
ITREKSVFDERDVAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMVRQALWLSNRDGHAVFEAALDATFRRHERLSQEQKTAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDKAVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDQSKTTLILAHLRRDVRTLNIMAREKLVERGIVGEGHVFKTADG
IRQFDAGDQIVFLKNEGSLGVKNGMIAHVIEAAPNRIIAVVGEGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGRLY
REALRFAESRGLNIVQVARTLVRDRLDWTLRQKTKLVDLGQRLAVFAERLGLHHPPKTQTMKEAAPMVAG
IKTFSGSVADTVENKLDTDPALKKQWEEVSARFAYVFADPETAFRAMNFDAVLTEKDTARQVVQKLEAEP
ASIGPLRGKTGILASKTEREDRRIAEINVPALKRDLEQYLQMREAAIKRLSTDEQVQRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTQRFGERTLLTNAAREPSGKLFDELSDG
MQPEQKERLKEAWPIMRTSQQLTAHERTVQTLRQAEDLRLSQRQTPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16033 | GenBank | WP_333722946 |
| Name | t4cp2_G6M86_RS29165_pQ15_94_4 |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 91814.81 Da Isoelectric Point: 5.7996
>WP_333722946.1 conjugal transfer protein TrbE [Agrobacterium tumefaciens]
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAIRVPTGDYPTEAESHFPDPVTRAIDDERRVHFQTERGHFESRHALILTWRPPEPRRSGLTR
YIYSDVGSRSATYADKALESFLTSIREVEQYLANVVSIRRMMTRETPERGGFRVARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPMVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDAVRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPVAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYAGAQVFA
FDKGGSMLPLTLGLDGDHYQIGGDIGPSAGGEVKALAFCPLAELSTDGDRAWASEWIEMLVALQGVTMTP
DYRNAISRQLSLMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGPFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIIATAIPKREYYVVSPGGRRLFNM
ALGPVALSFVGASGRQDLKRIRSLHSEHGAAWPLYWLQQRGIAHAETLFSAE
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAIRVPTGDYPTEAESHFPDPVTRAIDDERRVHFQTERGHFESRHALILTWRPPEPRRSGLTR
YIYSDVGSRSATYADKALESFLTSIREVEQYLANVVSIRRMMTRETPERGGFRVARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPMVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDAVRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPVAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYAGAQVFA
FDKGGSMLPLTLGLDGDHYQIGGDIGPSAGGEVKALAFCPLAELSTDGDRAWASEWIEMLVALQGVTMTP
DYRNAISRQLSLMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGPFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIIATAIPKREYYVVSPGGRRLFNM
ALGPVALSFVGASGRQDLKRIRSLHSEHGAAWPLYWLQQRGIAHAETLFSAE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16034 | GenBank | WP_333722978 |
| Name | traG_G6M86_RS29375_pQ15_94_4 |
UniProt ID | _ |
| Length | 641 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 641 a.a. Molecular weight: 69025.05 Da Isoelectric Point: 9.3160
>WP_333722978.1 Ti-type conjugative transfer system protein TraG [Agrobacterium tumefaciens]
MTVSKLFLAVIPATTMIVVVVLMPGVEHWLAAFGKTAQAKLMLGRIGLALPYATAAAIGTICLFAANGAV
GIKAAGWGVVSGNGMAILIAVLREGVRLAGIAGDIPKGQSVLGYADPATMLGASTTFLAGVFALRVAIKG
NAAFAKAAPRRIGGKRAVHGEADWMKVQEAAKLFPDPGGIVVGERYRVDRDSVAAVSFRADDPSTWGAGG
KNPLLCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSSEVAPMVSEHRRMAGRKVIILD
PSASGVGFNALDWIGRHGSTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVYLSGHTDE
EDQTLRRVRKNLSEPEPQLRARLTKIYEQSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGDSFSTDDLAKGETDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGAVNGRALFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSRQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRREDMKACVG
TNRFHREDATP
MTVSKLFLAVIPATTMIVVVVLMPGVEHWLAAFGKTAQAKLMLGRIGLALPYATAAAIGTICLFAANGAV
GIKAAGWGVVSGNGMAILIAVLREGVRLAGIAGDIPKGQSVLGYADPATMLGASTTFLAGVFALRVAIKG
NAAFAKAAPRRIGGKRAVHGEADWMKVQEAAKLFPDPGGIVVGERYRVDRDSVAAVSFRADDPSTWGAGG
KNPLLCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSSEVAPMVSEHRRMAGRKVIILD
PSASGVGFNALDWIGRHGSTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVYLSGHTDE
EDQTLRRVRKNLSEPEPQLRARLTKIYEQSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGDSFSTDDLAKGETDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGAVNGRALFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSRQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRREDMKACVG
TNRFHREDATP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 78068..96736
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| G6M86_RS29130 (G6M86_29050) | 73452..74666 | - | 1215 | WP_333722940 | plasmid replication protein RepC | - |
| G6M86_RS29135 (G6M86_29055) | 74821..75807 | - | 987 | WP_333722941 | plasmid partitioning protein RepB | - |
| G6M86_RS29140 (G6M86_29060) | 75882..77069 | - | 1188 | WP_071207840 | plasmid partitioning protein RepA | - |
| G6M86_RS29145 (G6M86_29065) | 77436..78071 | + | 636 | WP_333722943 | acyl-homoserine-lactone synthase | - |
| G6M86_RS29150 (G6M86_29070) | 78068..79039 | + | 972 | WP_333722944 | P-type conjugative transfer ATPase TrbB | virB11 |
| G6M86_RS29155 (G6M86_29075) | 79029..79433 | + | 405 | WP_333722945 | conjugal transfer pilin TrbC | virB2 |
| G6M86_RS29160 (G6M86_29080) | 79426..79725 | + | 300 | WP_080867871 | conjugal transfer protein TrbD | virB3 |
| G6M86_RS29165 (G6M86_29085) | 79735..82203 | + | 2469 | WP_333722946 | conjugal transfer protein TrbE | virb4 |
| G6M86_RS29170 (G6M86_29090) | 82217..82984 | + | 768 | WP_333723036 | P-type conjugative transfer protein TrbJ | virB5 |
| G6M86_RS29175 (G6M86_29095) | 82981..83208 | + | 228 | WP_333722947 | entry exclusion protein TrbK | - |
| G6M86_RS29180 (G6M86_29100) | 83202..84392 | + | 1191 | WP_333722948 | P-type conjugative transfer protein TrbL | virB6 |
| G6M86_RS29185 (G6M86_29105) | 84408..85085 | + | 678 | WP_333722949 | conjugal transfer protein TrbF | virB8 |
| G6M86_RS29190 (G6M86_29110) | 85103..85954 | + | 852 | WP_333723037 | P-type conjugative transfer protein TrbG | virB9 |
| G6M86_RS29195 (G6M86_29115) | 85954..86430 | + | 477 | WP_333722950 | conjugal transfer protein TrbH | - |
| G6M86_RS29200 (G6M86_29120) | 86442..87764 | + | 1323 | WP_333723038 | IncP-type conjugal transfer protein TrbI | virB10 |
| G6M86_RS29205 (G6M86_29125) | 87962..88123 | - | 162 | WP_333722951 | hypothetical protein | - |
| G6M86_RS29210 (G6M86_29130) | 88122..90974 | + | 2853 | WP_333722952 | hypothetical protein | - |
| G6M86_RS29215 (G6M86_29135) | 91093..92247 | - | 1155 | WP_333722953 | ROK family transcriptional regulator | - |
| G6M86_RS29220 (G6M86_29140) | 92439..93746 | + | 1308 | WP_333722954 | extracellular solute-binding protein | - |
| G6M86_RS29225 (G6M86_29145) | 93807..94697 | + | 891 | WP_333722955 | carbohydrate ABC transporter permease | - |
| G6M86_RS29230 (G6M86_29150) | 94701..95615 | + | 915 | WP_333722956 | ABC transporter permease subunit | - |
| G6M86_RS29235 (G6M86_29155) | 95618..96736 | + | 1119 | WP_333722957 | ABC transporter ATP-binding protein | virb4 |
| G6M86_RS29240 (G6M86_29160) | 96733..97590 | + | 858 | WP_333722958 | glycerophosphodiester phosphodiesterase family protein | - |
| G6M86_RS29245 (G6M86_29165) | 97624..98526 | + | 903 | WP_333722959 | inositol monophosphatase | - |
| G6M86_RS29250 (G6M86_29170) | 98711..100354 | + | 1644 | WP_333722960 | Na/Pi cotransporter family protein | - |
| G6M86_RS29255 (G6M86_29175) | 100396..101436 | - | 1041 | WP_333722961 | LacI family DNA-binding transcriptional regulator | - |
Host bacterium
| ID | 22167 | GenBank | NZ_CP049221 |
| Plasmid name | pQ15_94_4 | Incompatibility group | - |
| Plasmid size | 133442 bp | Coordinate of oriT [Strand] | 126850..126893 [-] |
| Host baterium | Agrobacterium tumefaciens strain Q15/94 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |