Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121662 |
| Name | oriT_pARO1.2 |
| Organism | Enterococcus faecalis ARO1/DG |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP022485 (55333..55382 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pARO1.2
AATATCGCAACATGGTACCATGTTGCTCTGCTCGCAAAAAGAAAGCCTAC
AATATCGCAACATGGTACCATGTTGCTCTGCTCGCAAAAAGAAAGCCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file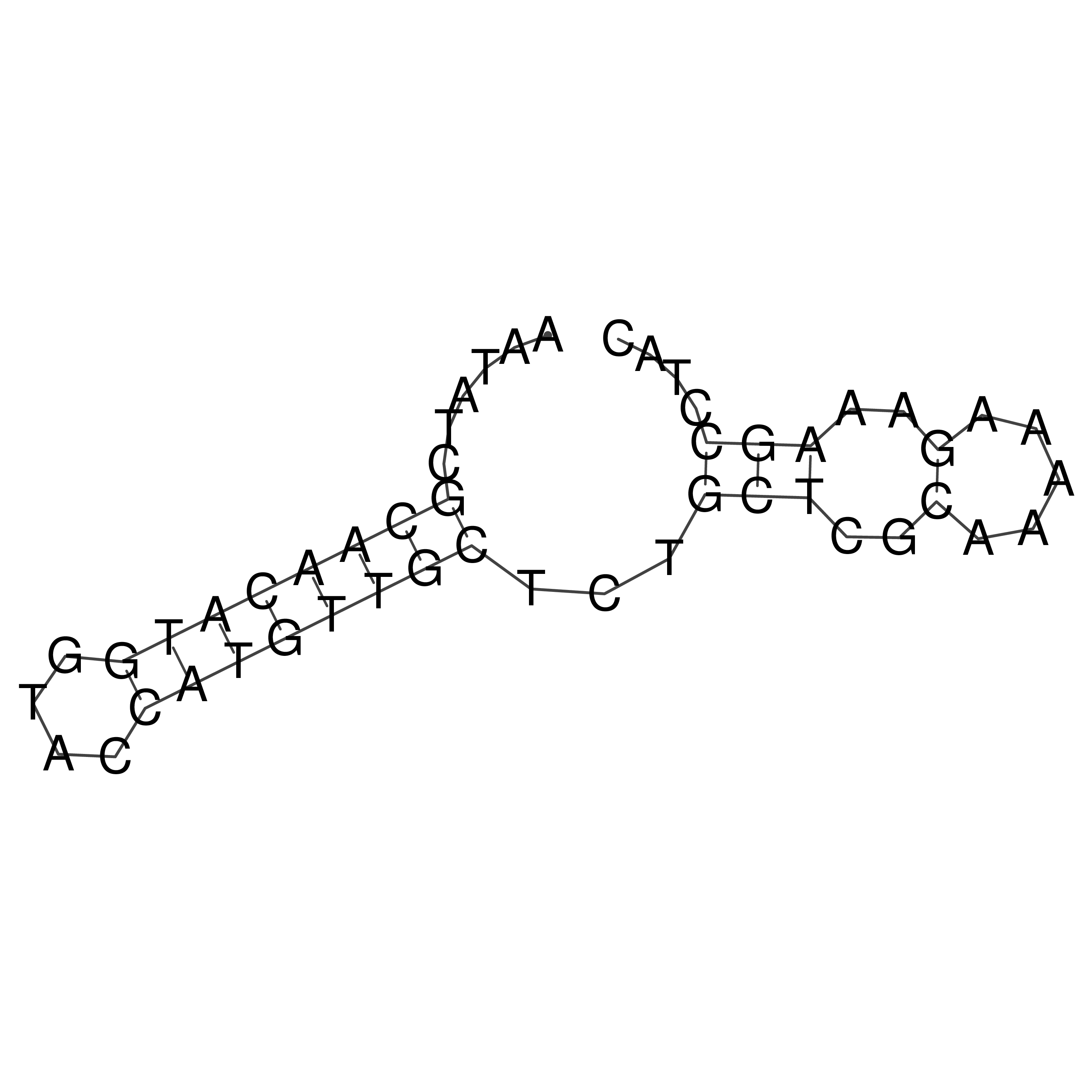
Relaxase
| ID | 13664 | GenBank | WP_002367697 |
| Name | Relaxase_CG806_RS00735_pARO1.2 |
UniProt ID | _ |
| Length | 561 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65315.04 Da Isoelectric Point: 6.1225
>WP_002367697.1 relaxase/mobilization nuclease domain-containing protein [Enterococcus faecalis]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKNS
HKKYTMWETESLYKEKIKSRLDFLLEQSSSINDFLEKAEALNLSVDFSKKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIVERLKENKSVLTLKEVVEKYGEKSEKSKNEFDYQLTIDEWQISHKTTRGYYVNVDFG
FGERGKVFIGAYKVDPLENGQYNIYVKRKDFFYLMNEKNADRNRYMTGETLIKQLRLYNGQTPLKKEPVM
QTIDELVNAINFLAANEIDDTRQLELLEEKLEAAFLEAEETLETLDEKMLELHQLSNLLLENELNGELEL
IQQKLKTLLPDATLAEFSYKDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKNS
HKKYTMWETESLYKEKIKSRLDFLLEQSSSINDFLEKAEALNLSVDFSKKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIVERLKENKSVLTLKEVVEKYGEKSEKSKNEFDYQLTIDEWQISHKTTRGYYVNVDFG
FGERGKVFIGAYKVDPLENGQYNIYVKRKDFFYLMNEKNADRNRYMTGETLIKQLRLYNGQTPLKKEPVM
QTIDELVNAINFLAANEIDDTRQLELLEEKLEAAFLEAEETLETLDEKMLELHQLSNLLLENELNGELEL
IQQKLKTLLPDATLAEFSYKDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7529 | GenBank | WP_002367696 |
| Name | WP_002367696_pARO1.2 |
UniProt ID | Q82YL4 |
| Length | 118 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 118 a.a. Molecular weight: 14042.10 Da Isoelectric Point: 9.9724
>WP_002367696.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Bacteria]
MENKQKLKRPIQRKIRFSKEENELINRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKEQS
MENKQKLKRPIQRKIRFSKEENELINRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKEQS
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q82YL4 |
T4CP
| ID | 15970 | GenBank | WP_002367691 |
| Name | t4cp2_CG806_RS00715_pARO1.2 |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 609 a.a. Molecular weight: 70145.14 Da Isoelectric Point: 6.1807
>WP_002367691.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecalis]
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRALEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRALEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 33951..52688
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CG806_RS00600 (CG806_00585) | 29134..29514 | + | 381 | WP_002367664 | hypothetical protein | - |
| CG806_RS00610 (CG806_00595) | 30085..30390 | + | 306 | WP_002367666 | hypothetical protein | - |
| CG806_RS00615 (CG806_00600) | 30401..33073 | + | 2673 | WP_002367667 | surface exclusion protein SEA1/PrgA | - |
| CG806_RS00620 (CG806_00605) | 33139..33726 | + | 588 | WP_002367669 | hypothetical protein | - |
| CG806_RS00625 (CG806_00610) | 33951..37865 | + | 3915 | WP_002367670 | LPXTG-anchored aggregation substance PrgB/Asc10 | prgB |
| CG806_RS00630 (CG806_00615) | 37927..38283 | + | 357 | WP_002367672 | pheromone response system RNA-binding regulator PrgU | - |
| CG806_RS00635 (CG806_00620) | 38311..39168 | + | 858 | WP_002367673 | pheromone response LPXTG-anchored adhesin PrgC | prgC |
| CG806_RS00640 (CG806_00625) | 39209..40108 | + | 900 | WP_025186739 | hypothetical protein | - |
| CG806_RS00645 (CG806_00630) | 40128..40562 | + | 435 | WP_002365857 | hypothetical protein | - |
| CG806_RS00650 (CG806_00635) | 40609..40836 | + | 228 | WP_002365858 | hypothetical protein | - |
| CG806_RS00655 (CG806_00640) | 40849..41145 | + | 297 | WP_002367677 | hypothetical protein | - |
| CG806_RS00660 (CG806_00645) | 41160..41960 | + | 801 | WP_002367679 | type IV secretion system protein | prgHb |
| CG806_RS00665 (CG806_00650) | 41962..42315 | + | 354 | WP_002365862 | PrgI family mobile element protein | prgIc |
| CG806_RS00670 (CG806_00655) | 42269..44659 | + | 2391 | WP_002365863 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| CG806_RS00675 (CG806_00660) | 44671..47286 | + | 2616 | WP_002367681 | phage tail tip lysozyme | prgK |
| CG806_RS00680 (CG806_00665) | 47310..47936 | + | 627 | WP_002367683 | hypothetical protein | prgL |
| CG806_RS00685 (CG806_00670) | 47923..48168 | + | 246 | WP_002365866 | hypothetical protein | - |
| CG806_RS00690 (CG806_00675) | 48152..48760 | + | 609 | WP_002367686 | hypothetical protein | - |
| CG806_RS00695 (CG806_00680) | 48874..49629 | + | 756 | WP_002367687 | Fic family protein | - |
| CG806_RS00700 (CG806_00685) | 49646..50104 | + | 459 | WP_002367688 | single-stranded DNA-binding protein | - |
| CG806_RS00705 (CG806_00690) | 50150..50332 | + | 183 | WP_002367689 | hypothetical protein | - |
| CG806_RS00710 (CG806_00695) | 50374..50859 | + | 486 | WP_002386493 | PcfB family protein | gbs1365 |
| CG806_RS00715 (CG806_00700) | 50859..52688 | + | 1830 | WP_002367691 | VirD4-like conjugal transfer protein, CD1115 family | virb4 |
| CG806_RS00720 (CG806_00705) | 52739..54898 | + | 2160 | WP_002367693 | ArdC-like ssDNA-binding domain-containing protein | - |
| CG806_RS00725 (CG806_00710) | 54931..55203 | + | 273 | WP_002367695 | LtrD | - |
| CG806_RS00730 (CG806_00715) | 55449..55805 | + | 357 | WP_002367696 | plasmid mobilization relaxosome protein MobC | - |
| CG806_RS00735 (CG806_00720) | 55806..57491 | + | 1686 | WP_002367697 | relaxase/mobilization nuclease domain-containing protein | - |
Host bacterium
| ID | 22089 | GenBank | NZ_CP022485 |
| Plasmid name | pARO1.2 | Incompatibility group | - |
| Plasmid size | 59380 bp | Coordinate of oriT [Strand] | 55333..55382 [+] |
| Host baterium | Enterococcus faecalis ARO1/DG |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | prgB/asc10 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |