Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121655 |
| Name | oriT_2022CK-00501|unnamed1 |
| Organism | Klebsiella variicola strain 2022CK-00501 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP114317 (202023..202072 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_2022CK-00501|unnamed1
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file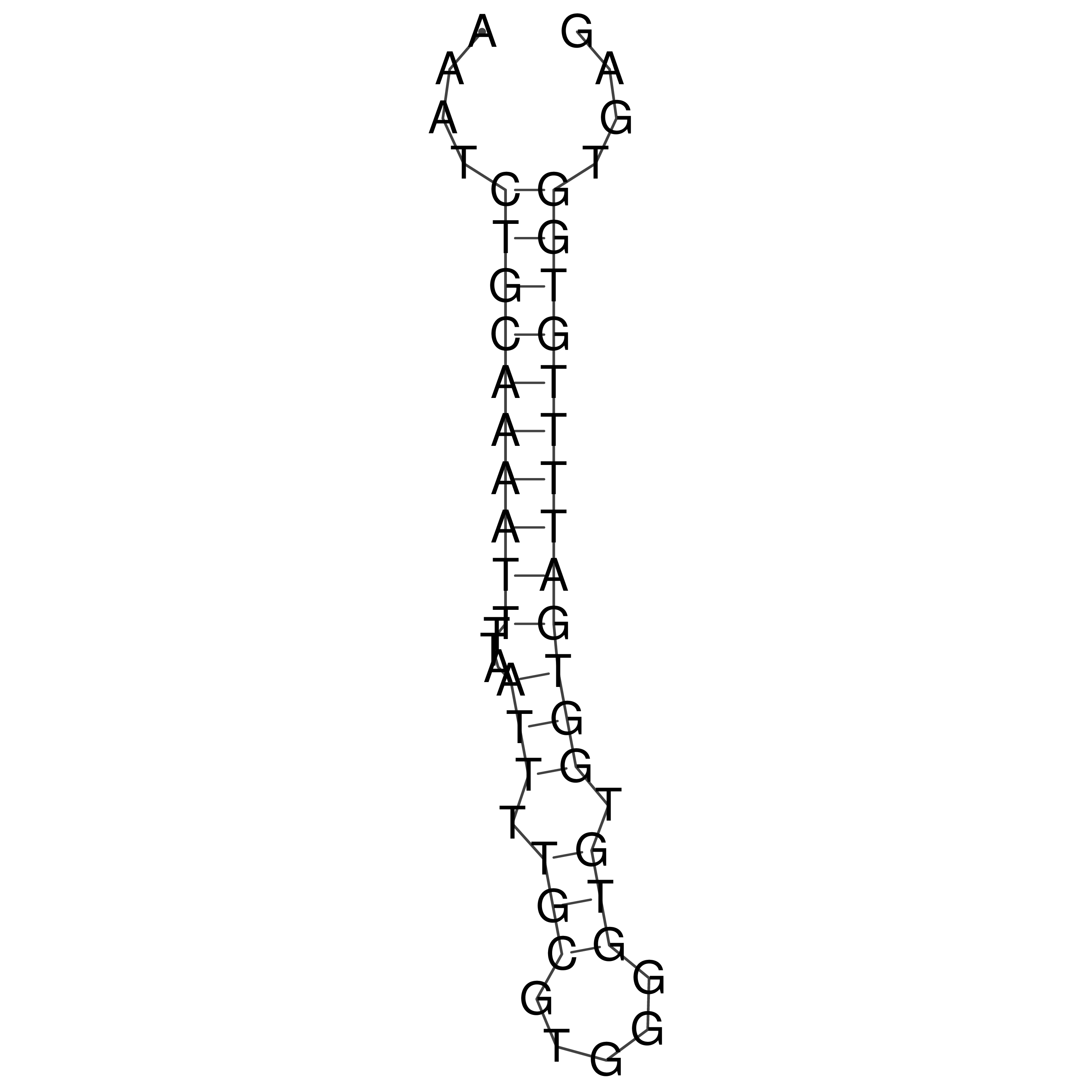
Relaxase
| ID | 13660 | GenBank | WP_269136661 |
| Name | traI_OGU15_RS27780_2022CK-00501|unnamed1 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190450.44 Da Isoelectric Point: 6.2888
>WP_269136661.1 conjugative transfer relaxase/helicase TraI [Klebsiella variicola]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAHGHGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPAQEKAIQ
KALSEGETLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAHGHGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPAQEKAIQ
KALSEGETLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15964 | GenBank | WP_095835928 |
| Name | traD_OGU15_RS27785_2022CK-00501|unnamed1 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85922.99 Da Isoelectric Point: 5.1149
>WP_095835928.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADITVSPAPVKAPPTTKMPAEEPSVRSTEPSALRLTTVPLIKPKAAAAAAAAATVSSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDNVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADITVSPAPVKAPPTTKMPAEEPSVRSTEPSALRLTTVPLIKPKAAAAAAAAATVSSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDNVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 172831..199525
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| OGU15_RS27785 (OGU15_27780) | 172831..175143 | - | 2313 | WP_095835928 | type IV conjugative transfer system coupling protein TraD | virb4 |
| OGU15_RS27790 (OGU15_27785) | 175503..176234 | - | 732 | WP_032415736 | conjugal transfer complement resistance protein TraT | - |
| OGU15_RS27795 (OGU15_27790) | 176486..177088 | - | 603 | WP_023280878 | hypothetical protein | - |
| OGU15_RS27800 (OGU15_27795) | 177222..178202 | + | 981 | WP_000019445 | IS5-like element ISKpn26 family transposase | - |
| OGU15_RS27805 (OGU15_27800) | 178303..181134 | - | 2832 | WP_269136662 | conjugal transfer mating-pair stabilization protein TraG | traG |
| OGU15_RS27810 (OGU15_27805) | 181134..182513 | - | 1380 | WP_011977731 | conjugal transfer pilus assembly protein TraH | traH |
| OGU15_RS27815 (OGU15_27810) | 182491..182934 | - | 444 | WP_015065638 | F-type conjugal transfer protein TrbF | - |
| OGU15_RS27820 (OGU15_27815) | 182979..183521 | - | 543 | WP_013054815 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| OGU15_RS27825 (OGU15_27820) | 183508..183735 | - | 228 | WP_114506536 | type-F conjugative transfer system pilin chaperone TraQ | - |
| OGU15_RS27830 (OGU15_27825) | 183746..184498 | - | 753 | WP_015065637 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| OGU15_RS27835 (OGU15_27830) | 184519..184845 | - | 327 | WP_032436744 | hypothetical protein | - |
| OGU15_RS29220 | 184891..185076 | - | 186 | WP_032436745 | hypothetical protein | - |
| OGU15_RS27840 (OGU15_27835) | 185073..185309 | - | 237 | WP_032436746 | conjugal transfer protein TrbE | - |
| OGU15_RS27845 (OGU15_27840) | 185341..187296 | - | 1956 | WP_023307537 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| OGU15_RS27850 (OGU15_27845) | 187355..187993 | - | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| OGU15_RS27855 (OGU15_27850) | 188006..188965 | - | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| OGU15_RS27860 (OGU15_27855) | 189009..189635 | - | 627 | WP_023307538 | type-F conjugative transfer system protein TraW | traW |
| OGU15_RS27865 (OGU15_27860) | 189635..190024 | - | 390 | WP_004152591 | type-F conjugative transfer system protein TrbI | - |
| OGU15_RS27870 (OGU15_27865) | 190024..192663 | - | 2640 | WP_016528865 | type IV secretion system protein TraC | virb4 |
| OGU15_RS27875 (OGU15_27870) | 192735..193133 | - | 399 | WP_072158599 | hypothetical protein | - |
| OGU15_RS27880 (OGU15_27875) | 193141..193431 | - | 291 | WP_023307542 | hypothetical protein | - |
| OGU15_RS27885 (OGU15_27880) | 193428..193832 | - | 405 | WP_023307543 | hypothetical protein | - |
| OGU15_RS27890 (OGU15_27885) | 193899..194216 | - | 318 | WP_023307544 | hypothetical protein | - |
| OGU15_RS27895 (OGU15_27890) | 194217..194435 | - | 219 | WP_004171484 | hypothetical protein | - |
| OGU15_RS27900 (OGU15_27895) | 194459..194749 | - | 291 | WP_032423381 | hypothetical protein | - |
| OGU15_RS27905 (OGU15_27900) | 194754..195164 | - | 411 | WP_269136663 | hypothetical protein | - |
| OGU15_RS27910 (OGU15_27905) | 195296..195865 | - | 570 | WP_023307547 | type IV conjugative transfer system lipoprotein TraV | traV |
| OGU15_RS27915 (OGU15_27910) | 196077..196490 | - | 414 | Protein_205 | conjugal transfer pilus-stabilizing protein TraP | - |
| OGU15_RS27920 (OGU15_27915) | 196483..197907 | - | 1425 | WP_023307549 | F-type conjugal transfer pilus assembly protein TraB | traB |
| OGU15_RS27925 (OGU15_27920) | 197907..198647 | - | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| OGU15_RS27930 (OGU15_27925) | 198634..199200 | - | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| OGU15_RS27935 (OGU15_27930) | 199220..199525 | - | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| OGU15_RS27940 (OGU15_27935) | 199539..199907 | - | 369 | WP_023307550 | type IV conjugative transfer system pilin TraA | - |
| OGU15_RS27945 (OGU15_27940) | 199961..200332 | - | 372 | WP_004208838 | TraY domain-containing protein | - |
| OGU15_RS27950 (OGU15_27945) | 200416..201111 | - | 696 | WP_023307551 | transcriptional regulator TraJ family protein | - |
| OGU15_RS27955 (OGU15_27950) | 201318..201710 | - | 393 | WP_004206766 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| OGU15_RS27960 (OGU15_27955) | 202217..202624 | + | 408 | WP_020805750 | transglycosylase SLT domain-containing protein | - |
| OGU15_RS27965 (OGU15_27960) | 202662..203192 | - | 531 | WP_015632491 | antirestriction protein | - |
Host bacterium
| ID | 22082 | GenBank | NZ_CP114317 |
| Plasmid name | 2022CK-00501|unnamed1 | Incompatibility group | IncFIB |
| Plasmid size | 236561 bp | Coordinate of oriT [Strand] | 202023..202072 [+] |
| Host baterium | Klebsiella variicola strain 2022CK-00501 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE, arsC, arsB, arsA, arsD, arsR, arsH |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |