Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121600 |
| Name | oriT_pT212 |
| Organism | Proteus mirabilis strain T21 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP017084 (75628..75778 [-], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 93..98, 106..111 (TGGCCT..AGGCCA) 12..17, 24..29 (AACCCT..AGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_pT212
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file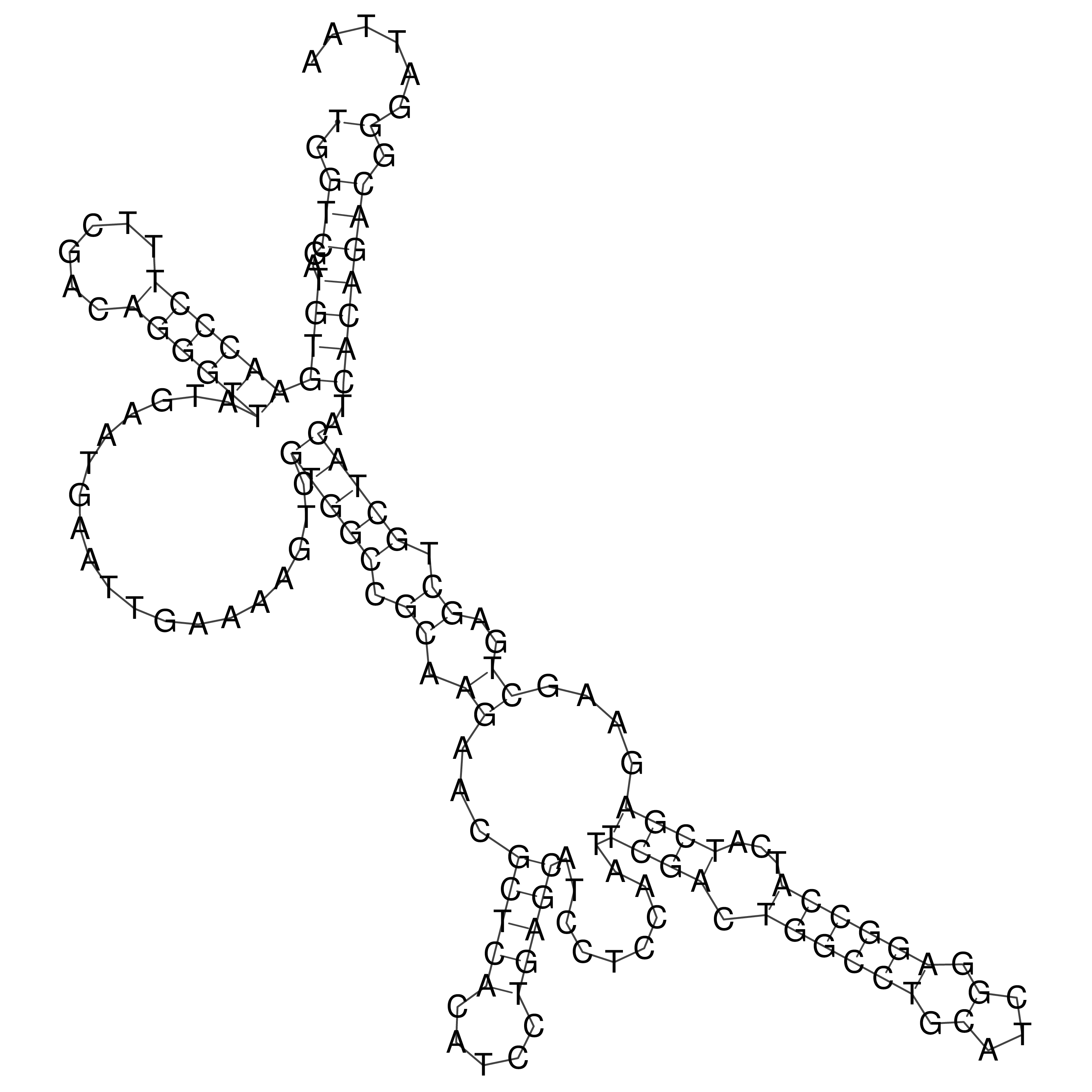
Relaxase
| ID | 13621 | GenBank | WP_001337757 |
| Name | mobH_BG257_RS19910_pT212 |
UniProt ID | A0A709FRI2 |
| Length | 992 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 992 a.a. Molecular weight: 109581.85 Da Isoelectric Point: 4.8956
>WP_001337757.1 MULTISPECIES: MobH family relaxase [Gammaproteobacteria]
MLKALNKLFGGRSGVIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEADDGDDQEEGEAALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGPTESSK
PDAGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPF
DAFNASAETTSTDATNSEIPDVAMPGKQEEQPKQDFVPQEQNSLQGDDFPMFGGSDEPPSWAIEPLPMLT
DAPEQPTHTPEMPHTDNVNQHEKDAKTLLVEMLSGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVI
LYPDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQ
DAFELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKASPEQKAKGKDSQPQPKEKKVDVTSPVEEQQ
RKPVQEKQNVARLPKREAQPVAPEPKVEREKELGHVEVREREDPEVREFEPPKAKTNPKDINAEDFLPSG
VTPQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKK
RQGKLYLEVNET
MLKALNKLFGGRSGVIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEADDGDDQEEGEAALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGPTESSK
PDAGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPF
DAFNASAETTSTDATNSEIPDVAMPGKQEEQPKQDFVPQEQNSLQGDDFPMFGGSDEPPSWAIEPLPMLT
DAPEQPTHTPEMPHTDNVNQHEKDAKTLLVEMLSGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVI
LYPDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQ
DAFELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKASPEQKAKGKDSQPQPKEKKVDVTSPVEEQQ
RKPVQEKQNVARLPKREAQPVAPEPKVEREKELGHVEVREREDPEVREFEPPKAKTNPKDINAEDFLPSG
VTPQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKK
RQGKLYLEVNET
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A709FRI2 |
T4CP
| ID | 15921 | GenBank | WP_000637386 |
| Name | traC_BG257_RS19830_pT212 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 92590.87 Da Isoelectric Point: 5.9222
>WP_000637386.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTERIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRQQAA
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTERIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRQQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15922 | GenBank | WP_000178856 |
| Name | traD_BG257_RS19905_pT212 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69197.66 Da Isoelectric Point: 6.9403
>WP_000178856.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 52503..77694
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BG257_RS19770 (BG257_19370) | 47630..48538 | - | 909 | WP_000739139 | hypothetical protein | - |
| BG257_RS19775 (BG257_19375) | 48549..49517 | - | 969 | WP_000085162 | AAA family ATPase | - |
| BG257_RS19780 (BG257_19380) | 49737..49922 | - | 186 | WP_001186917 | hypothetical protein | - |
| BG257_RS19785 (BG257_19385) | 50226..50546 | - | 321 | WP_000547566 | hypothetical protein | - |
| BG257_RS19790 (BG257_19390) | 50840..51481 | + | 642 | WP_000796664 | hypothetical protein | - |
| BG257_RS19795 (BG257_19395) | 51604..52464 | + | 861 | WP_000709517 | hypothetical protein | - |
| BG257_RS19800 (BG257_19400) | 52503..55301 | - | 2799 | WP_001256487 | conjugal transfer mating pair stabilization protein TraN | traN |
| BG257_RS19805 (BG257_19405) | 55417..56421 | - | 1005 | WP_043940352 | TraU family protein | traU |
| BG257_RS19810 (BG257_19410) | 56421..57092 | - | 672 | WP_001337754 | EAL domain-containing protein | - |
| BG257_RS19815 (BG257_19415) | 57089..58354 | - | 1266 | WP_000621288 | TrbC family F-type conjugative pilus assembly protein | traW |
| BG257_RS19820 (BG257_19420) | 58317..58847 | - | 531 | WP_001010738 | S26 family signal peptidase | - |
| BG257_RS19825 (BG257_19425) | 58844..59161 | - | 318 | WP_000351984 | hypothetical protein | - |
| BG257_RS19830 (BG257_19430) | 59176..61623 | - | 2448 | WP_000637386 | type IV secretion system protein TraC | virb4 |
| BG257_RS19835 (BG257_19435) | 61620..62327 | - | 708 | WP_001259347 | DsbC family protein | - |
| BG257_RS19840 (BG257_19440) | 62476..68007 | - | 5532 | WP_085281193 | Ig-like domain-containing protein | - |
| BG257_RS19845 (BG257_19445) | 68208..68600 | - | 393 | WP_000479535 | TraA family conjugative transfer protein | - |
| BG257_RS19850 (BG257_19450) | 68604..69182 | - | 579 | WP_000793435 | type IV conjugative transfer system lipoprotein TraV | traV |
| BG257_RS19855 (BG257_19455) | 69179..70492 | - | 1314 | WP_024131605 | TraB/VirB10 family protein | traB |
| BG257_RS19860 (BG257_19460) | 70492..71409 | - | 918 | WP_096043119 | type-F conjugative transfer system secretin TraK | traK |
| BG257_RS19865 (BG257_19465) | 71393..72019 | - | 627 | WP_001049716 | TraE/TraK family type IV conjugative transfer system protein | traE |
| BG257_RS19870 (BG257_19470) | 72016..72297 | - | 282 | WP_000805625 | type IV conjugative transfer system protein TraL | traL |
| BG257_RS19875 (BG257_19475) | 72442..72807 | - | 366 | WP_001052531 | hypothetical protein | - |
| BG257_RS20960 | 72819..72962 | - | 144 | WP_001275801 | hypothetical protein | - |
| BG257_RS19880 (BG257_19480) | 73143..73805 | - | 663 | WP_001231463 | hypothetical protein | - |
| BG257_RS19885 (BG257_19485) | 73805..74182 | - | 378 | WP_000869261 | hypothetical protein | - |
| BG257_RS20965 | 74424..74600 | - | 177 | WP_169342285 | hypothetical protein | - |
| BG257_RS19895 (BG257_19490) | 74648..75277 | - | 630 | WP_000743450 | DUF4400 domain-containing protein | tfc7 |
| BG257_RS19900 (BG257_19495) | 75234..75779 | - | 546 | WP_000228720 | hypothetical protein | - |
| BG257_RS19905 (BG257_19500) | 75829..77694 | - | 1866 | WP_000178856 | conjugative transfer system coupling protein TraD | virb4 |
| BG257_RS19910 (BG257_19505) | 77691..80669 | - | 2979 | WP_001337757 | MobH family relaxase | - |
| BG257_RS19915 (BG257_19510) | 80837..81454 | + | 618 | WP_001249396 | hypothetical protein | - |
| BG257_RS19920 (BG257_19515) | 81436..81669 | + | 234 | WP_001191892 | hypothetical protein | - |
Host bacterium
| ID | 22027 | GenBank | NZ_CP017084 |
| Plasmid name | pT212 | Incompatibility group | IncA/C2 |
| Plasmid size | 171489 bp | Coordinate of oriT [Strand] | 75628..75778 [-] |
| Host baterium | Proteus mirabilis strain T21 |
Cargo genes
| Drug resistance gene | blaCTX-M-14, sul2, aph(3'')-Ib, aph(6)-Id, blaTEM-1B, aac(3)-IId, dfrA12, aadA2, aph(3')-Ia, rmtB, tet(G) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |