Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121529 |
| Name | oriT_Sym1|unnamed1 |
| Organism | Roseibium sp. Sym1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP114787 (57560..57594 [+], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 19..25, 29..35 (CGTCGCG..CGCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
>oriT_Sym1|unnamed1
TCCAAGGGCGCAGTAATACGTCGCGAACCGCGACG
TCCAAGGGCGCAGTAATACGTCGCGAACCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file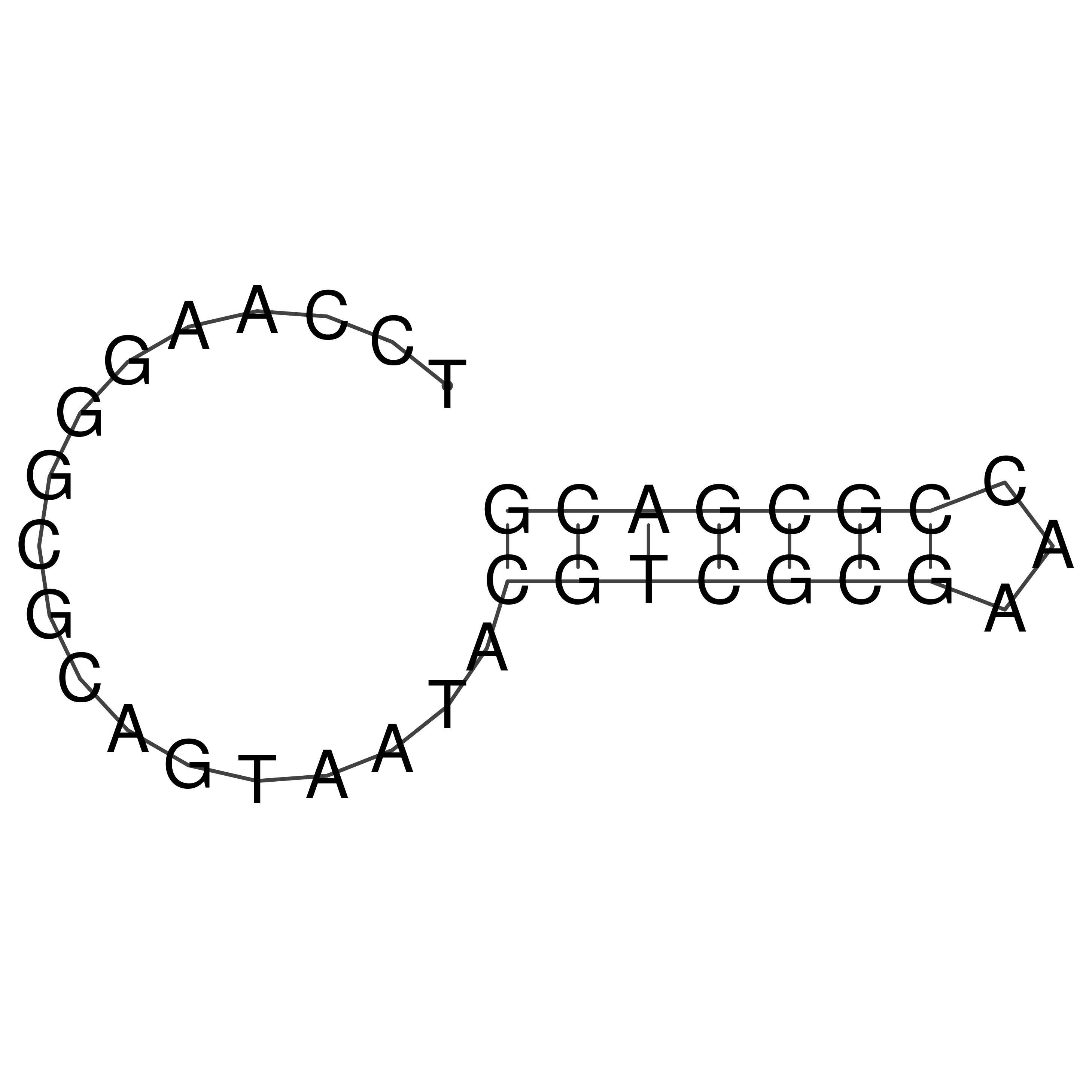
Relaxase
| ID | 13573 | GenBank | WP_269586448 |
| Name | traA_O6760_RS30895_Sym1|unnamed1 |
UniProt ID | _ |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122894.06 Da Isoelectric Point: 10.0124
>WP_269586448.1 Ti-type conjugative transfer relaxase TraA [Roseibium sp. Sym1]
MAITHFTPQLISRGNGRSAVLSAAYRHCAKMSHEAEARTVDYSNKNNLAHEEFLLPADAPKWARDMIADH
SVAGAAEAFWNAVEAFEKRSDAQLAKEFIIALPVELSRDQNIALVRQFVSEQVLARGQVADWVLHHDPGN
PHIHLMTTLRPLTEDGFGSKKVAVMGPDGQPVRMKTGKIQYKLWSGEKPEFLEQRQRWLDLQNHHLALAG
LEIRVDGRSYAERGIDIEPTTHIGVAAKALQRKAETKAGAIDLERLALHEAQRQKNAARIEVRPELVLDI
LTSEKSVFDQRDIAKVLHRYIDDPESFQRLLARIAESPECLKLADETIDFATGARVPARLTTRAMIRLEA
EMVSRADWLSGKSGFAVRGKVLEAVFGRHERLSEEQRTAIEHVAGSERIATVVGRAGAGKTTMMKAAREA
WELAGYTVVGGALAGKAAEGLEKEAGIPSRTLASWELGWQKGRRSLDDRTVFVLDEAGMVSSRQMATFVE
AVTRSGAKLILVGDAEQLQPIEAGAAFRAIVERTGYAELETIYRQNEPWMRAASLDLARGRVKEALSSYR
AKGKLIAQTLKADAIETLIADWNRDYDPEKSALILAHLRRDVRQLNTMAREKLVERGIIEEGYAFRTEDG
PRQFSSGDQIVFLKNEGSLGVKNGMIGKVVEAAPGRIVAMIGNGEGATRVTVDQRFYRNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDMALYYGSRSFHLAGGLEKLLSRKNAKEVTLDYAGGRFY
AQALRFANNRGLHLVRVARTLMHDRVQWTIRQKQKLVDLGSRLRAAGARLGLAHRPLQQLPQSIRRAEPM
VKGISNFTMSVTDATDEKLQSDPALQKQWDNFSDRIRLVYEDPESAFKAMRMEAVVEDPNVARLRLGEIE
RSAASFGALKGREGLLASRADREDRRVAEVNVPALRKDLERYLDMRERATAKYRLEEEGHRKRTSIDIPS
LSPAAARAMEKVRDAIDRNDLPAALGFALTDRMVKAELDTFNKAVSERFGERTLLGNAAKTPAGATFDKA
AQGMSPGDREKLSTAWPTMRAGQQLAAHERTQQALKQSEAMWQAQTKSQRLKQ
MAITHFTPQLISRGNGRSAVLSAAYRHCAKMSHEAEARTVDYSNKNNLAHEEFLLPADAPKWARDMIADH
SVAGAAEAFWNAVEAFEKRSDAQLAKEFIIALPVELSRDQNIALVRQFVSEQVLARGQVADWVLHHDPGN
PHIHLMTTLRPLTEDGFGSKKVAVMGPDGQPVRMKTGKIQYKLWSGEKPEFLEQRQRWLDLQNHHLALAG
LEIRVDGRSYAERGIDIEPTTHIGVAAKALQRKAETKAGAIDLERLALHEAQRQKNAARIEVRPELVLDI
LTSEKSVFDQRDIAKVLHRYIDDPESFQRLLARIAESPECLKLADETIDFATGARVPARLTTRAMIRLEA
EMVSRADWLSGKSGFAVRGKVLEAVFGRHERLSEEQRTAIEHVAGSERIATVVGRAGAGKTTMMKAAREA
WELAGYTVVGGALAGKAAEGLEKEAGIPSRTLASWELGWQKGRRSLDDRTVFVLDEAGMVSSRQMATFVE
AVTRSGAKLILVGDAEQLQPIEAGAAFRAIVERTGYAELETIYRQNEPWMRAASLDLARGRVKEALSSYR
AKGKLIAQTLKADAIETLIADWNRDYDPEKSALILAHLRRDVRQLNTMAREKLVERGIIEEGYAFRTEDG
PRQFSSGDQIVFLKNEGSLGVKNGMIGKVVEAAPGRIVAMIGNGEGATRVTVDQRFYRNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDMALYYGSRSFHLAGGLEKLLSRKNAKEVTLDYAGGRFY
AQALRFANNRGLHLVRVARTLMHDRVQWTIRQKQKLVDLGSRLRAAGARLGLAHRPLQQLPQSIRRAEPM
VKGISNFTMSVTDATDEKLQSDPALQKQWDNFSDRIRLVYEDPESAFKAMRMEAVVEDPNVARLRLGEIE
RSAASFGALKGREGLLASRADREDRRVAEVNVPALRKDLERYLDMRERATAKYRLEEEGHRKRTSIDIPS
LSPAAARAMEKVRDAIDRNDLPAALGFALTDRMVKAELDTFNKAVSERFGERTLLGNAAKTPAGATFDKA
AQGMSPGDREKLSTAWPTMRAGQQLAAHERTQQALKQSEAMWQAQTKSQRLKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15859 | GenBank | WP_269586447 |
| Name | traG_O6760_RS30880_Sym1|unnamed1 |
UniProt ID | _ |
| Length | 640 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 640 a.a. Molecular weight: 69133.99 Da Isoelectric Point: 8.4545
>WP_269586447.1 Ti-type conjugative transfer system protein TraG [Roseibium sp. Sym1]
MTLNKLLLVIAPVGLMMAVCLGLTGSETWLAQFGNSPEAQRTLGQIGIALPYVSAAALGVLVLFAARGAM
AIKSAGLGVLVGAVGVIGIAGMREFSRLSGFADAVPDGHRLIEYADPSTGIGAMVCFIAGMFALRVALRG
DAAFASMKPRRLTGKRAIHGSADWMSLGEAKRMFGEDGGIVVGEAYRVDKDSVASHTFRADDKQSWGAGG
KSQLLCFDASFGSTHGLVFAGSGGFKTTSVTIPTALKWGGGLVALDPSNEVAPMVIDHRRKAGREVFVLS
PKDASVGFNVLDWIGRYGGTKEEDVVAVASWVVTDIGRQTSMRDDFFRASALQLITALIADVCLSGHTAK
ENQTLRQVRMNLSEPEPKLRERLQSIHDNSESDFVRENVAAFVNMTPETFSGVYANAIKETHWLSYPNYA
ALVSGSSLSTDDLANGETDIFVNLDLKTLEAHPGLARTIIGALLNAIYNRNGETKGRTLFLLDEVARLGY
LRILETARDAGRKYGISLLLLFQSIGQMREIYGGRDATSKWFESASWVSFAAINDPETAEYISRRCGNTT
VEVDQLSRTSQMSGSSRTRSKQLAARPLILPEEVLRMRGDEQIAFTSGNPPLRCGRAIWFRRGDMKSVVA
TNRFHREGQG
MTLNKLLLVIAPVGLMMAVCLGLTGSETWLAQFGNSPEAQRTLGQIGIALPYVSAAALGVLVLFAARGAM
AIKSAGLGVLVGAVGVIGIAGMREFSRLSGFADAVPDGHRLIEYADPSTGIGAMVCFIAGMFALRVALRG
DAAFASMKPRRLTGKRAIHGSADWMSLGEAKRMFGEDGGIVVGEAYRVDKDSVASHTFRADDKQSWGAGG
KSQLLCFDASFGSTHGLVFAGSGGFKTTSVTIPTALKWGGGLVALDPSNEVAPMVIDHRRKAGREVFVLS
PKDASVGFNVLDWIGRYGGTKEEDVVAVASWVVTDIGRQTSMRDDFFRASALQLITALIADVCLSGHTAK
ENQTLRQVRMNLSEPEPKLRERLQSIHDNSESDFVRENVAAFVNMTPETFSGVYANAIKETHWLSYPNYA
ALVSGSSLSTDDLANGETDIFVNLDLKTLEAHPGLARTIIGALLNAIYNRNGETKGRTLFLLDEVARLGY
LRILETARDAGRKYGISLLLLFQSIGQMREIYGGRDATSKWFESASWVSFAAINDPETAEYISRRCGNTT
VEVDQLSRTSQMSGSSRTRSKQLAARPLILPEEVLRMRGDEQIAFTSGNPPLRCGRAIWFRRGDMKSVVA
TNRFHREGQG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15860 | GenBank | WP_209181194 |
| Name | t4cp2_O6760_RS31495_Sym1|unnamed1 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 91050.42 Da Isoelectric Point: 5.3166
>WP_209181194.1 MULTISPECIES: conjugal transfer protein TrbE [Stappiaceae]
MVALKQFRNSKPSFSDLLPYAGLVADGVILLKDGSLMAGWYFAGPDSESSTNIERNEVSRQINAVLSRLG
SGWMIQVEAVRVPTTAYPAREDCHFPDPVTRAIDEQRRDHFMAERGHYESQHALILTYRPPEPRRSAMAR
YVYSDEGSRSQTYADGVLTSFQTSIREVEQYLGNVLMIRRMVTRTVVHPESGDEARYDELFQFIRFCIFG
GNYPVRLPAIPMYLDWLVTAEFHHGLSPMVDGSYLAVVGIDGFPAESWPGILNSLDLMPLTYRWSSRFIF
LDDQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAAASSQLVAYGYYTPVIVM
FDTDEARLRDQAEAVRRLIQAEGFGARIETLNATEAYLGSLPGNSYANVREPLINTRNLADLIPVNSVWA
GSAVAPCPFYPPNAPPLMQVASGSSPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGKSLYPLTRAASGDHYEIGAGEGTALAFCPLAELSTDGDRAWAAEWVETLVAMQGVTITPDHRNAIA
RQIGLLADARGRSLSDFVSGVQMREIKTALQHYTVDGPMGQLLDAEEDGLTLGSFQTFEVEELMNMGERN
LVPVLLYLFRRIEKRLTGSPSLIVLDEAWLMLGHPMFRDKIREWLKVLRKANCAVVLATQSISDAEQSGI
IDVLKESCPTKICLPNGAARESGTREFYERIGFNDRQIEIVATAIPKREYYVASPEGRRLFDMSLGPVAL
AFVGASGKDNLKRIDQLSEAHGEEWPAAWLEERGVDDAKRILTHS
MVALKQFRNSKPSFSDLLPYAGLVADGVILLKDGSLMAGWYFAGPDSESSTNIERNEVSRQINAVLSRLG
SGWMIQVEAVRVPTTAYPAREDCHFPDPVTRAIDEQRRDHFMAERGHYESQHALILTYRPPEPRRSAMAR
YVYSDEGSRSQTYADGVLTSFQTSIREVEQYLGNVLMIRRMVTRTVVHPESGDEARYDELFQFIRFCIFG
GNYPVRLPAIPMYLDWLVTAEFHHGLSPMVDGSYLAVVGIDGFPAESWPGILNSLDLMPLTYRWSSRFIF
LDDQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAAASSQLVAYGYYTPVIVM
FDTDEARLRDQAEAVRRLIQAEGFGARIETLNATEAYLGSLPGNSYANVREPLINTRNLADLIPVNSVWA
GSAVAPCPFYPPNAPPLMQVASGSSPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGKSLYPLTRAASGDHYEIGAGEGTALAFCPLAELSTDGDRAWAAEWVETLVAMQGVTITPDHRNAIA
RQIGLLADARGRSLSDFVSGVQMREIKTALQHYTVDGPMGQLLDAEEDGLTLGSFQTFEVEELMNMGERN
LVPVLLYLFRRIEKRLTGSPSLIVLDEAWLMLGHPMFRDKIREWLKVLRKANCAVVLATQSISDAEQSGI
IDVLKESCPTKICLPNGAARESGTREFYERIGFNDRQIEIVATAIPKREYYVASPEGRRLFDMSLGPVAL
AFVGASGKDNLKRIDQLSEAHGEEWPAAWLEERGVDDAKRILTHS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 170433..179972
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| O6760_RS31425 | 165944..166783 | + | 840 | WP_209181243 | MaoC family dehydratase N-terminal domain-containing protein | - |
| O6760_RS31430 | 166777..167568 | + | 792 | WP_209181202 | CoA ester lyase | - |
| O6760_RS31435 | 167760..168298 | - | 539 | Protein_156 | IS5 family transposase | - |
| O6760_RS31440 | 168334..168941 | - | 608 | Protein_157 | transposase | - |
| O6760_RS31445 | 169003..169431 | + | 429 | WP_269586463 | helix-turn-helix domain-containing protein | - |
| O6760_RS31450 | 169386..169594 | - | 209 | Protein_159 | GMP synthase (glutamine-hydrolyzing) | - |
| O6760_RS31455 | 169657..170361 | - | 705 | WP_209181200 | autoinducer binding domain-containing protein | - |
| O6760_RS31460 | 170433..171737 | - | 1305 | WP_209181199 | IncP-type conjugal transfer protein TrbI | virB10 |
| O6760_RS31465 | 171734..172201 | - | 468 | WP_209181198 | conjugal transfer protein TrbH | - |
| O6760_RS31470 | 172206..172988 | - | 783 | WP_269586464 | P-type conjugative transfer protein TrbG | virB9 |
| O6760_RS31475 | 173107..173769 | - | 663 | WP_209181241 | conjugal transfer protein TrbF | virB8 |
| O6760_RS31480 | 173778..174944 | - | 1167 | WP_209181196 | P-type conjugative transfer protein TrbL | virB6 |
| O6760_RS31485 | 174925..175128 | - | 204 | WP_209181195 | hypothetical protein | - |
| O6760_RS31490 | 175125..175922 | - | 798 | WP_209181240 | P-type conjugative transfer protein TrbJ | virB5 |
| O6760_RS31495 | 175894..178341 | - | 2448 | WP_209181194 | conjugal transfer protein TrbE | virb4 |
| O6760_RS31500 | 178348..178647 | - | 300 | WP_209181193 | conjugal transfer protein TrbD | virB3 |
| O6760_RS31505 | 178640..179011 | - | 372 | WP_209181192 | TrbC/VirB2 family protein | virB2 |
| O6760_RS31510 | 179001..179972 | - | 972 | WP_209181191 | P-type conjugative transfer ATPase TrbB | virB11 |
| O6760_RS31515 | 180491..181702 | + | 1212 | WP_209181239 | plasmid partitioning protein RepA | - |
| O6760_RS31520 | 181911..182915 | + | 1005 | WP_209181190 | plasmid partitioning protein RepB | - |
| O6760_RS31525 | 183109..184413 | + | 1305 | WP_209181189 | plasmid replication protein RepC | - |
| O6760_RS31530 | 184442..184657 | - | 216 | WP_209181188 | hypothetical protein | - |
Host bacterium
| ID | 21956 | GenBank | NZ_CP114787 |
| Plasmid name | Sym1|unnamed1 | Incompatibility group | - |
| Plasmid size | 192102 bp | Coordinate of oriT [Strand] | 57560..57594 [+] |
| Host baterium | Roseibium sp. Sym1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | ncrA |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | - |