Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121513 |
| Name | oriT_FDAARGOS_909|unnamed |
| Organism | Delftia acidovorans strain FDAARGOS_909 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP065667 (72216..72312 [+], 97 nt) |
| oriT length | 97 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 97 nt
GAATAAGGGACAGCGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCGCCTATCCTGCCCGCCTTACGGCGTTGATAACACCAAGGAAAGTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file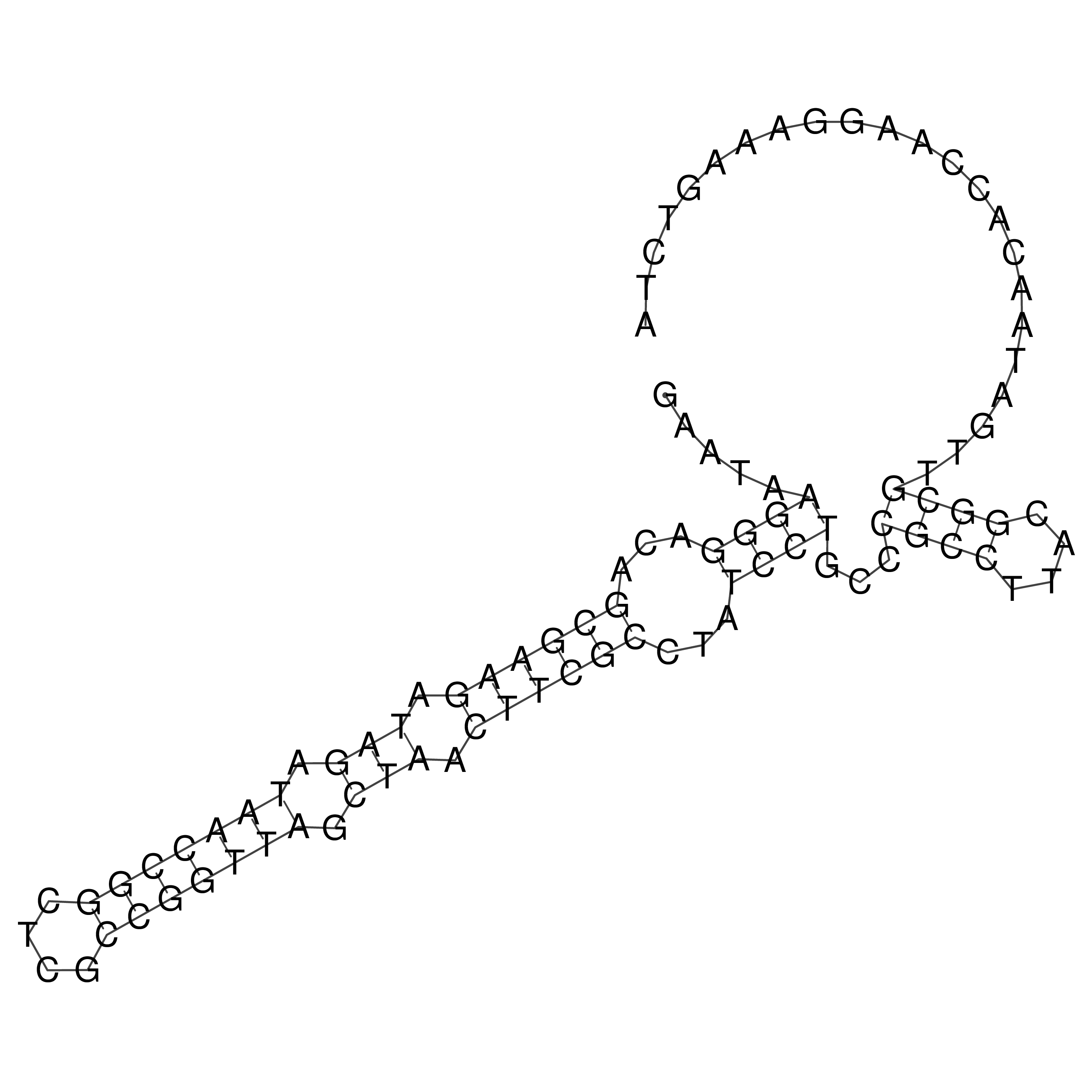
Relaxase
| ID | 13563 | GenBank | WP_011798731 |
| Name | traI_I6G66_RS00395_FDAARGOS_909|unnamed |
UniProt ID | A0A7T2VWK8 |
| Length | 752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 752 a.a. Molecular weight: 83050.85 Da Isoelectric Point: 10.8263
MIVKHVPMRSLGKSDFAGLVNYVTDAQSKDHRLGQVQITNCDAMSVQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGFAEHQRVSAVHNDTDNLHIHIAINKIHPTRHTMHEPFGQHRT
LAELCTALERDYGLERDNHEPRQRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRERANGLVIEAGDGTMVKASTLARDLSKPKLEARLGPFEPSPERQTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRGQALEQARRRKDRLVEAAKRSNRLRRATIKVVGESRTNKKLLYAQASKALRDE
IQAINKQYQQDRQALYDSHSRRTWADWLKKEATHGNAEALAALRAREAAQGLQGNTIQGRGDARPGHAPV
TDNITKKGTIIFRAGLSAVRDDGDKLQVSREATREGLQEALRLAMERYGSRITVNGTTQFKAQVIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTERADQRPAEHRGRAGRGPGGPGQRAAAHDDAARPVAIA
RAGFGRPGAGRTDAAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRDVPRHL
EQQGTQPDHQLRRGISRPGTGVATAPPGLAAADKYIAEREAKRSKGFDIPKHARYTDGDGALAFQGVRNI
EGQALALLKRGDEVMVMPIDQATARRLSRIAVGDAVTITPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7T2VWK8 |
Auxiliary protein
| ID | 7461 | GenBank | WP_011013290 |
| Name | WP_011013290_FDAARGOS_909|unnamed |
UniProt ID | A0A7T2VWC8 |
| Length | 123 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 123 a.a. Molecular weight: 13936.01 Da Isoelectric Point: 8.4443
MEADEQPTAKRRQHLRVPVFPEEKELIEANAKRAGVSVARYLRDVGQGYQIKGVMDYEYVRELVRVNGDL
GRLGGLLKLWLTDDPRTARFGDATILALLGRIEATQDEMSRLMKSVVQPRAEP
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7T2VWC8 |
T4CP
| ID | 15848 | GenBank | WP_011798706 |
| Name | t4cp2_I6G66_RS00245_FDAARGOS_909|unnamed |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 93902.02 Da Isoelectric Point: 6.4729
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSSAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKLT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILTLNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDAAIREAIRQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15849 | GenBank | WP_011798733 |
| Name | t4cp2_I6G66_RS00385_FDAARGOS_909|unnamed |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69613.65 Da Isoelectric Point: 8.5000
MKIKMNNAVGPQVRSAKAKTPKLLPILGGVSLVGGLQAATQFFAYTFQYHASLGANLGHVYAPWSILNWS
AKWYSQYPDEIMRAGSIGMVTATVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKDAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSSGGVCWNPLDEIRVGTEAEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKARNEGGTATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKDLMHSDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFQARAAIPAPKATDRLRQVVEAGE
GITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 40223..50908
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| I6G66_RS00205 (I6G66_00205) | 36420..37223 | + | 804 | WP_088138647 | IS21-like element ISThsp10 family helper ATPase IstB | - |
| I6G66_RS00210 (I6G66_00210) | 37114..37641 | + | 528 | WP_256943238 | Tn3 family transposase | - |
| I6G66_RS00215 (I6G66_00215) | 37832..39055 | - | 1224 | WP_088138646 | plasmid replication initiator TrfA | - |
| I6G66_RS00220 (I6G66_00220) | 39103..39441 | - | 339 | WP_011798708 | single-stranded DNA-binding protein | - |
| I6G66_RS00225 (I6G66_00225) | 39550..39912 | + | 363 | WP_000283041 | transcriptional regulator | - |
| I6G66_RS00230 (I6G66_00230) | 40223..41188 | + | 966 | WP_011798707 | P-type conjugative transfer ATPase TrbB | virB11 |
| I6G66_RS00235 (I6G66_00235) | 41201..41665 | + | 465 | WP_011222079 | conjugal transfer system pilin TrbC | virB2 |
| I6G66_RS00240 (I6G66_00240) | 41669..41980 | + | 312 | WP_001207961 | conjugal transfer protein TrbD | virB3 |
| I6G66_RS00245 (I6G66_00245) | 41977..44535 | + | 2559 | WP_011798706 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| I6G66_RS00250 (I6G66_00250) | 44532..45314 | + | 783 | WP_011798705 | conjugal transfer protein TrbF | virB8 |
| I6G66_RS00255 (I6G66_00255) | 45332..46231 | + | 900 | WP_000753228 | P-type conjugative transfer protein TrbG | virB9 |
| I6G66_RS00260 (I6G66_00260) | 46234..46722 | + | 489 | WP_011798704 | conjugal transfer protein TrbH | - |
| I6G66_RS00265 (I6G66_00265) | 46727..48145 | + | 1419 | WP_011798703 | TrbI/VirB10 family protein | virB10 |
| I6G66_RS00270 (I6G66_00270) | 48163..48927 | + | 765 | WP_011798702 | P-type conjugative transfer protein TrbJ | virB5 |
| I6G66_RS00275 (I6G66_00275) | 48937..49164 | + | 228 | WP_011798701 | entry exclusion lipoprotein TrbK | - |
| I6G66_RS00280 (I6G66_00280) | 49175..50908 | + | 1734 | WP_011798700 | P-type conjugative transfer protein TrbL | virB6 |
| I6G66_RS00285 (I6G66_00285) | 50925..51509 | + | 585 | WP_011798699 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| I6G66_RS00290 (I6G66_00290) | 51523..52158 | + | 636 | WP_011798698 | transglycosylase SLT domain-containing protein | - |
| I6G66_RS00295 (I6G66_00295) | 52187..52453 | + | 267 | WP_011798697 | hypothetical protein | - |
| I6G66_RS00300 (I6G66_00300) | 52453..53151 | + | 699 | WP_011798696 | TraX family protein | - |
| I6G66_RS00305 (I6G66_00305) | 53167..53598 | + | 432 | WP_001665065 | OmpA family protein | - |
| I6G66_RS00310 (I6G66_00310) | 53754..54428 | + | 675 | WP_011798695 | class I SAM-dependent methyltransferase | - |
| I6G66_RS00315 (I6G66_00315) | 54507..55037 | + | 531 | WP_011798694 | hypothetical protein | - |
| I6G66_RS00320 (I6G66_00320) | 54973..55191 | - | 219 | Protein_63 | helix-turn-helix domain-containing protein | - |
| I6G66_RS00325 (I6G66_00325) | 55206..55439 | + | 234 | WP_011798693 | hypothetical protein | - |
| I6G66_RS00330 (I6G66_00330) | 55503..55709 | - | 207 | WP_011798692 | hypothetical protein | - |
| I6G66_RS00335 (I6G66_00335) | 55706..55879 | - | 174 | WP_172734609 | hypothetical protein | - |
Host bacterium
| ID | 21940 | GenBank | NZ_CP065667 |
| Plasmid name | FDAARGOS_909|unnamed | Incompatibility group | - |
| Plasmid size | 73470 bp | Coordinate of oriT [Strand] | 72216..72312 [+] |
| Host baterium | Delftia acidovorans strain FDAARGOS_909 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF24 |