Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121511 |
| Name | oriT_pE1532-NDM |
| Organism | Enterobacter hormaechei strain E1532 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP114575 (48998..49615 [+], 618 nt) |
| oriT length | 618 nt |
| IRs (inverted repeats) | 589..594, 608..613 (TCCATG..CATGGA) 592..597, 602..607 (ATGCCC..GGGCAT) 520..527, 530..537 (CCTCCCGT..ACGGGAGG) 455..460, 464..469 (ATTCGC..GCGAAT) 197..202, 208..213 (TTCAAT..ATTGAA) 85..90, 96..101 (TGCTTT..AAAGCA) |
| Location of nic site | 315..316 |
| Conserved sequence flanking the nic site |
TCCTGCATCG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 618 nt
>oriT_pE1532-NDM
TTCTTTGTTTCCCTGTCAAATACATTGTGGTTTTGATAAAATCATCATTGTCTTTTGACACATTAACATCTTCATTTTTAACCTTGCTTTTCAGTAAAGCATTGGCAAGCAGCTTCATTCCTTTTGCCACGGTTAAACCCGGTTCATCTGGATAAAGTGTTTTAAGACAATCAAGTAGATCATCATGTTGGCTTTCTTCAATCCTGAATTGAACTTTTTTCATAAAACTCTCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGTTGCGGAATGGCGTCAGAGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAACTACCTCCACACATAAATTGTGCTTCAGCGTCCATCGAAATGCAAAAAATGAAAGGCTATCGTGGCTGTGATCTGGTGCGTCTGCGACCGGAGACAACGGATTCGCGGCTAACTTGGGCGGAAATAAATTCGCTACGCGAATGGCCTGGCAGGGGGCGCGAGCGCTGTTTTACGGAATATACAAAAAAAGCACCTCCCGTAAACGGGAGGGCTTCGGCGATTCAGGAACGGGAATTTTATTCTGCCTCTGGTGTGTCGCCTTCCATGCCCTGCCGGGCATCATGGAGCCAG
TTCTTTGTTTCCCTGTCAAATACATTGTGGTTTTGATAAAATCATCATTGTCTTTTGACACATTAACATCTTCATTTTTAACCTTGCTTTTCAGTAAAGCATTGGCAAGCAGCTTCATTCCTTTTGCCACGGTTAAACCCGGTTCATCTGGATAAAGTGTTTTAAGACAATCAAGTAGATCATCATGTTGGCTTTCTTCAATCCTGAATTGAACTTTTTTCATAAAACTCTCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGTTGCGGAATGGCGTCAGAGACGTCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAACTACCTCCACACATAAATTGTGCTTCAGCGTCCATCGAAATGCAAAAAATGAAAGGCTATCGTGGCTGTGATCTGGTGCGTCTGCGACCGGAGACAACGGATTCGCGGCTAACTTGGGCGGAAATAAATTCGCTACGCGAATGGCCTGGCAGGGGGCGCGAGCGCTGTTTTACGGAATATACAAAAAAAGCACCTCCCGTAAACGGGAGGGCTTCGGCGATTCAGGAACGGGAATTTTATTCTGCCTCTGGTGTGTCGCCTTCCATGCCCTGCCGGGCATCATGGAGCCAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file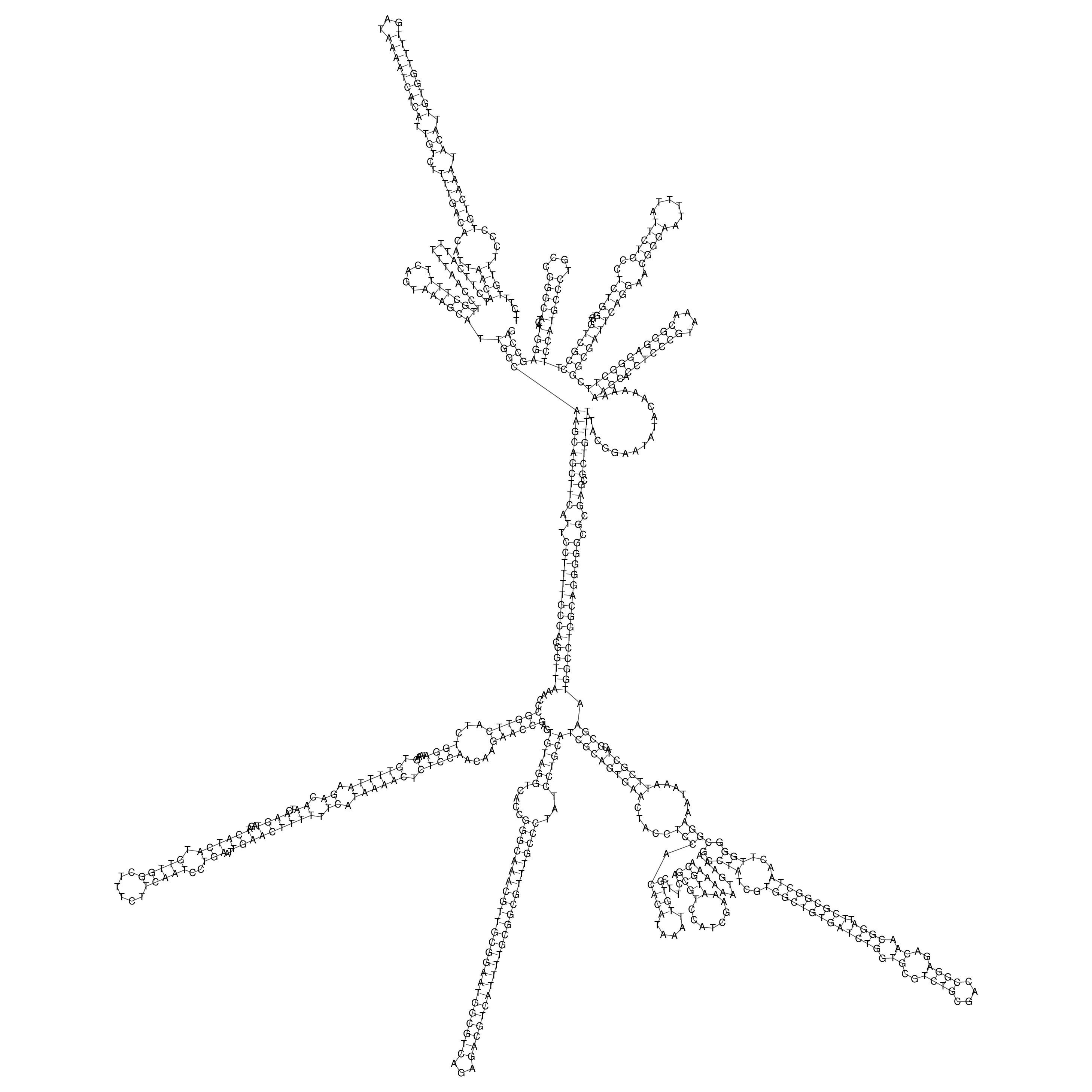
Relaxase
| ID | 13561 | GenBank | WP_004199101 |
| Name | mobP1_O2601_RS26295_pE1532-NDM |
UniProt ID | A0A515HL38 |
| Length | 386 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 386 a.a. Molecular weight: 44388.82 Da Isoelectric Point: 10.6017
>WP_004199101.1 MULTISPECIES: MobP1 family relaxase [Enterobacterales]
MGVYVDKEFRVKRKSSEAGRKSAFAHKVKNGGKNYQRNVQERINRKGASKEVVVKISGGAITRQGVRNSI
DYMSRESELPVMSESGQVWKGDEIQEAKEHMIDRANDPQNVFDDKGKENKKVTQNIVFSPPVSAKVKPED
LLESVRKTMNKKYPNHRFVLGYHNDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKMKGYD
VKATHKQQHGLNQSIKDAHKTAPKRQKGVYEVVDVGYDHYQNDKTKPKQHFIKLKTLNKGVEKTYWGADF
GELTTRENVKKGDLVKLKKLGQKEVKIPALDKNGVQHGWKTAHRNEWQLENLGVKGIDRISSSSKELVLN
SAEMIKKQQLQMRNFSQIKQSMIQSEQKVKIGIRLG
MGVYVDKEFRVKRKSSEAGRKSAFAHKVKNGGKNYQRNVQERINRKGASKEVVVKISGGAITRQGVRNSI
DYMSRESELPVMSESGQVWKGDEIQEAKEHMIDRANDPQNVFDDKGKENKKVTQNIVFSPPVSAKVKPED
LLESVRKTMNKKYPNHRFVLGYHNDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKMKGYD
VKATHKQQHGLNQSIKDAHKTAPKRQKGVYEVVDVGYDHYQNDKTKPKQHFIKLKTLNKGVEKTYWGADF
GELTTRENVKKGDLVKLKKLGQKEVKIPALDKNGVQHGWKTAHRNEWQLENLGVKGIDRISSSSKELVLN
SAEMIKKQQLQMRNFSQIKQSMIQSEQKVKIGIRLG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A515HL38 |
T4CP
| ID | 15846 | GenBank | WP_000053822 |
| Name | t4cp2_O2601_RS26225_pE1532-NDM |
UniProt ID | _ |
| Length | 611 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 611 a.a. Molecular weight: 69204.04 Da Isoelectric Point: 7.4342
>WP_000053822.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacterales]
MSLKLPDKGQWAFIGLVMCLVTYYTGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFIIWQLNKTDVALYGDAKFASDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREKLLGNKVYLLDPFNSKTHQFNPLF
YIDLKEESGAKDLLKLIEILFPSYGLTGAEAHFNNLAGQYWTGLAKLLHFFINYDPSWLGEFGLKPVFSI
GSVVDLYSNIDRELILSKREDLEGTKGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDMSLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGVEGAKTLMSAHPCRIIYAVSEEDDAKKIS
EKLGYITTKSTGSSKTSGRSTSKSSSESEAQRALVLPQELGTLDFKEEFIILKGENPVKAEKALYFLDPY
FMDRLMMVSPKLTELTASLNKTKKVLGVKGLKYPSKEKMLSVGELESEVLL
MSLKLPDKGQWAFIGLVMCLVTYYTGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFIIWQLNKTDVALYGDAKFASDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREKLLGNKVYLLDPFNSKTHQFNPLF
YIDLKEESGAKDLLKLIEILFPSYGLTGAEAHFNNLAGQYWTGLAKLLHFFINYDPSWLGEFGLKPVFSI
GSVVDLYSNIDRELILSKREDLEGTKGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDMSLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGVEGAKTLMSAHPCRIIYAVSEEDDAKKIS
EKLGYITTKSTGSSKTSGRSTSKSSSESEAQRALVLPQELGTLDFKEEFIILKGENPVKAEKALYFLDPY
FMDRLMMVSPKLTELTASLNKTKKVLGVKGLKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15847 | GenBank | WP_015060057 |
| Name | t4cp2_O2601_RS26270_pE1532-NDM |
UniProt ID | _ |
| Length | 917 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 917 a.a. Molecular weight: 104788.72 Da Isoelectric Point: 6.2666
>WP_015060057.1 MULTISPECIES: VirB3 family type IV secretion system protein [Enterobacterales]
MSTVFKGLTRPALIRGLGVPLYPFLGMCVICVLLGVWIHEAMYALILPGWYAIKRVTKIDERFFDLLYLR
MQIKGNPLANKRFNAVHYAGSSYDAVDISKVDNFMKLKDQSSLEELIPYSSHITDNLIVTRNHDLLATWQ
IDGAYFECVDEADLALLTDQLNTLIRSFDGKPVTFYTHRIRVRKEVRPVFDSKIPFVNRVMNDYYESLSA
AEYFENKLYLTVCYKPFSAEDKVTHFLSRKKGNKNIFEEPINDMNEICGRLSTYLSRFHSRRLGLYEENN
IVYSEQLTLFQKLLSGRWQKVRVTNSPFYTYLGGKDLFFGNDAGQITASDHARYFRCIEIKDYFQETDAG
IFDALMYLPVEYVQTSSLTPIDKQSAIKALDDQIDKLEMTDDAAKSLLADLKVGLDMVSSGYISFGKSHQ
TLIVYADSPERLVKDTNIVTTTLEDLGLIVTYSTLSLGAAYFAQLPGNYTLRPRLSSISSLNFAEMESFH
NFFTGKEKGNTWGNNLITLGGSGNDIYNLNYHMTTEHQNYFGKNPTLGHTEILGTSNVGKTVVMMTKAFA
AQQFGTPESFPPERKLKKLTTVFFDKDRAAEPGIRSMGGAYFRVKEGEPTGWNPAALPPTKRNISFMKDL
VRLLCTLNSEPLDDYQNRLISDAVERLMQRSNRSYPISKLRPLILEPDDTETRRHGLKARLNAWVQGGEF
GWVFDNPDDTFDVDNLDVFGIDGTEFLDNKVLSSAASFYLIYRVTMLADGRRLLIYMDEFWQWINNEAFR
DFVYNKLKTARKLDMVLVVATQSPDELIKSPIAAAVREQCATHIYLANPKAKRSEYVDELEVRELYFDKI
KAIDPLSRQFLVVKNPQRKGESDDFAAFARLDLGKAAYYLPVLSASKPQLELFDEIWKEGMKPEEWLDTY
LERANLI
MSTVFKGLTRPALIRGLGVPLYPFLGMCVICVLLGVWIHEAMYALILPGWYAIKRVTKIDERFFDLLYLR
MQIKGNPLANKRFNAVHYAGSSYDAVDISKVDNFMKLKDQSSLEELIPYSSHITDNLIVTRNHDLLATWQ
IDGAYFECVDEADLALLTDQLNTLIRSFDGKPVTFYTHRIRVRKEVRPVFDSKIPFVNRVMNDYYESLSA
AEYFENKLYLTVCYKPFSAEDKVTHFLSRKKGNKNIFEEPINDMNEICGRLSTYLSRFHSRRLGLYEENN
IVYSEQLTLFQKLLSGRWQKVRVTNSPFYTYLGGKDLFFGNDAGQITASDHARYFRCIEIKDYFQETDAG
IFDALMYLPVEYVQTSSLTPIDKQSAIKALDDQIDKLEMTDDAAKSLLADLKVGLDMVSSGYISFGKSHQ
TLIVYADSPERLVKDTNIVTTTLEDLGLIVTYSTLSLGAAYFAQLPGNYTLRPRLSSISSLNFAEMESFH
NFFTGKEKGNTWGNNLITLGGSGNDIYNLNYHMTTEHQNYFGKNPTLGHTEILGTSNVGKTVVMMTKAFA
AQQFGTPESFPPERKLKKLTTVFFDKDRAAEPGIRSMGGAYFRVKEGEPTGWNPAALPPTKRNISFMKDL
VRLLCTLNSEPLDDYQNRLISDAVERLMQRSNRSYPISKLRPLILEPDDTETRRHGLKARLNAWVQGGEF
GWVFDNPDDTFDVDNLDVFGIDGTEFLDNKVLSSAASFYLIYRVTMLADGRRLLIYMDEFWQWINNEAFR
DFVYNKLKTARKLDMVLVVATQSPDELIKSPIAAAVREQCATHIYLANPKAKRSEYVDELEVRELYFDKI
KAIDPLSRQFLVVKNPQRKGESDDFAAFARLDLGKAAYYLPVLSASKPQLELFDEIWKEGMKPEEWLDTY
LERANLI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 36308..46417
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| O2601_RS26200 (O2601_26200) | 31431..32693 | - | 1263 | WP_000517490 | ATP-binding protein | - |
| O2601_RS26205 (O2601_26205) | 32776..33270 | - | 495 | WP_001215543 | thermonuclease family protein | - |
| O2601_RS26210 (O2601_26210) | 33267..33593 | - | 327 | WP_004208538 | hypothetical protein | - |
| O2601_RS26215 (O2601_26215) | 33679..33993 | - | 315 | WP_000754496 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| O2601_RS26220 (O2601_26220) | 34081..34473 | - | 393 | WP_000738524 | cag pathogenicity island Cag12 family protein | - |
| O2601_RS26225 (O2601_26225) | 34470..36305 | - | 1836 | WP_000053822 | type IV secretory system conjugative DNA transfer family protein | - |
| O2601_RS26230 (O2601_26230) | 36308..37342 | - | 1035 | WP_000650512 | P-type DNA transfer ATPase VirB11 | virB11 |
| O2601_RS26235 (O2601_26235) | 37339..37536 | - | 198 | WP_000629109 | DNA-binding protein | - |
| O2601_RS26240 (O2601_26240) | 37538..38752 | - | 1215 | WP_001295060 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| O2601_RS26245 (O2601_26245) | 38749..39678 | - | 930 | WP_000783386 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| O2601_RS26250 (O2601_26250) | 39684..40412 | - | 729 | WP_004199085 | virB8 family protein | virB8 |
| O2601_RS26255 (O2601_26255) | 40607..41662 | - | 1056 | WP_000235886 | type IV secretion system protein | virB6 |
| O2601_RS26260 (O2601_26260) | 41674..41931 | - | 258 | WP_000759841 | EexN family lipoprotein | - |
| O2601_RS26265 (O2601_26265) | 41941..42711 | - | 771 | WP_000716326 | type IV secretion system protein | - |
| O2601_RS26270 (O2601_26270) | 42721..45474 | - | 2754 | WP_015060057 | VirB3 family type IV secretion system protein | virb4 |
| O2601_RS26275 (O2601_26275) | 45499..45789 | - | 291 | WP_000916156 | TrbC/VirB2 family protein | virB2 |
| O2601_RS26280 (O2601_26280) | 45773..46417 | - | 645 | WP_000953538 | lytic transglycosylase domain-containing protein | virB1 |
| O2601_RS26285 (O2601_26285) | 46463..46699 | - | 237 | WP_001348696 | hypothetical protein | - |
| O2601_RS26290 (O2601_26290) | 46650..47159 | - | 510 | WP_000455437 | transcription termination/antitermination NusG family protein | - |
| O2601_RS26295 (O2601_26295) | 47512..48672 | - | 1161 | WP_004199101 | MobP1 family relaxase | - |
| O2601_RS26300 (O2601_26300) | 48675..49220 | - | 546 | WP_001348698 | DNA distortion polypeptide 1 | - |
| O2601_RS26305 (O2601_26305) | 49561..49818 | - | 258 | WP_004199355 | hypothetical protein | - |
| O2601_RS26310 (O2601_26310) | 49815..49979 | - | 165 | WP_000004083 | hypothetical protein | - |
| O2601_RS26315 (O2601_26315) | 50054..50392 | - | 339 | WP_001099725 | hypothetical protein | - |
| O2601_RS26320 (O2601_26320) | 50486..50728 | - | 243 | WP_000008707 | hypothetical protein | - |
| O2601_RS26325 (O2601_26325) | 50718..50987 | - | 270 | WP_001006717 | hypothetical protein | - |
Host bacterium
| ID | 21938 | GenBank | NZ_CP114575 |
| Plasmid name | pE1532-NDM | Incompatibility group | IncX3 |
| Plasmid size | 53769 bp | Coordinate of oriT [Strand] | 48998..49615 [+] |
| Host baterium | Enterobacter hormaechei strain E1532 |
Cargo genes
| Drug resistance gene | blaSHV-12, blaNDM-1 |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |