Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121500 |
| Name | oriT_SRS16|5 |
| Organism | Variovorax sp. SRS16 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LR594670 (6794..6888 [-], 95 nt) |
| oriT length | 95 nt |
| IRs (inverted repeats) | 26..32, 37..43 (TAACCGG..CCGGTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 95 nt
>oriT_SRS16|5
CGAATAAGGGGATAGTGAAGATAGATAACCGGCTTTCCGGTTAGCTAACTTCACCTATCCTGCCCGCCTTTCGGCGGTATGAACACCAAGGAAAG
CGAATAAGGGGATAGTGAAGATAGATAACCGGCTTTCCGGTTAGCTAACTTCACCTATCCTGCCCGCCTTTCGGCGGTATGAACACCAAGGAAAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file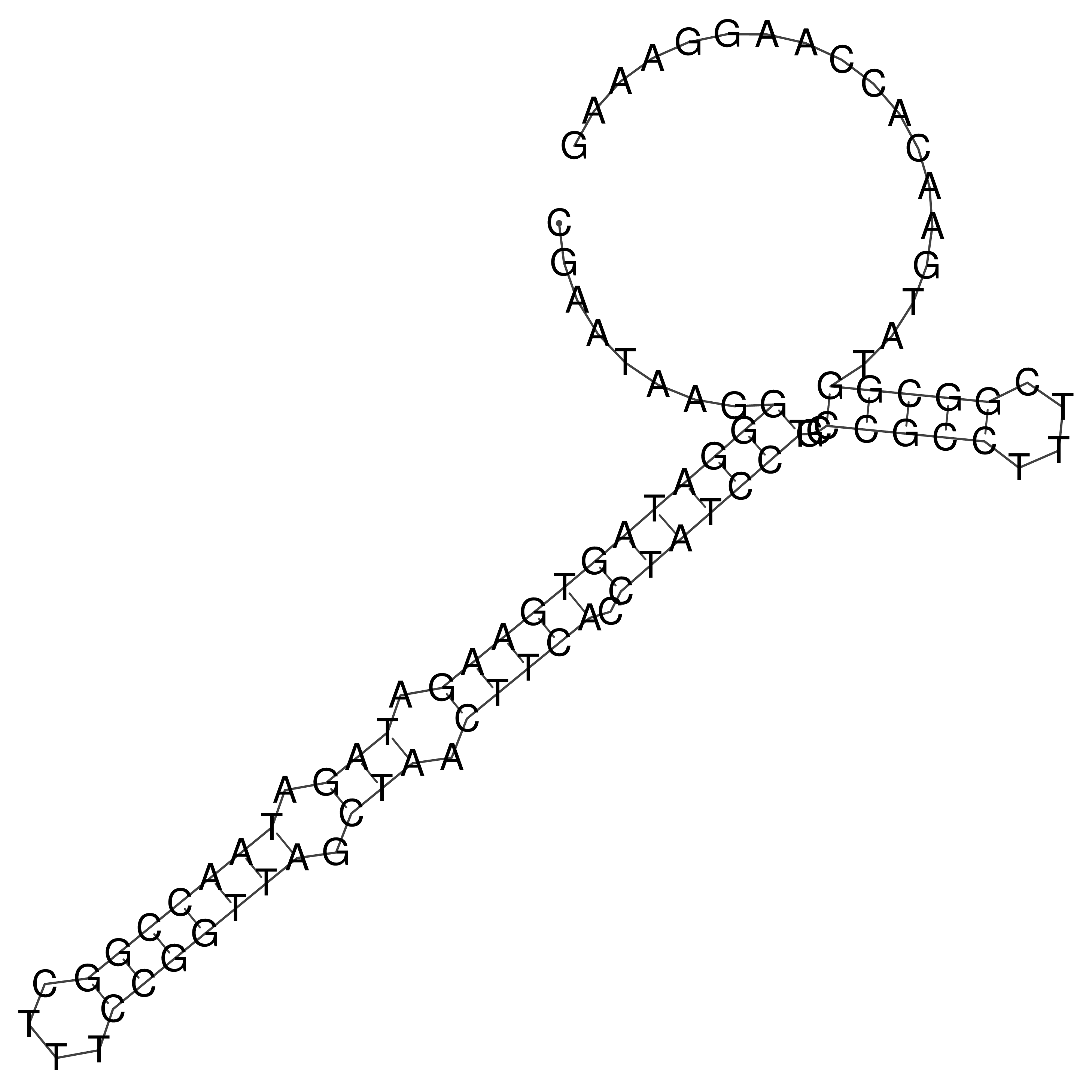
Relaxase
| ID | 13555 | GenBank | WP_015063533 |
| Name | traI_E5P2_RS37340_SRS16|5 |
UniProt ID | K4IYE5 |
| Length | 730 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 730 a.a. Molecular weight: 80015.48 Da Isoelectric Point: 10.3609
>WP_015063533.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Burkholderiales]
MIAKHVSMKTVQKSDFAGLVKYITDEQAKNERVGYVAVTNCHTDRPEVAITEVLNTQAQNTRSGADKTYH
LIVSFRPGEQPDDATLKAIEARICEGLGFGEHQRVSAVHHDTDNLHIHIAINKIHPTRYTILEPFNAYHT
LGQLCEKLEREYGLERDNHQGNKLVSEGRAQDMERHAGVESLQGWVKRECAADMQAAQSWEALHQVMRDN
GLELRERGNGFVIQAADGLAVKASSIGREFSKAKLEERLGTFEPSAEHLERAAQKPRKQYDKKPVRSRVN
TVELHAKYKADQLTISGSRTAEWEKARAKKDRLIEAAKRSGRLKRSTIKLISGAGVGKKLMYSATSKTMK
SEIEKINKQYLKERQEIHEKYQRRAWADWLRAKAAEGDQEALGALRARDAAQGLKGNTVAGTGPLRPQAA
AAEQDSVTKKGTIIYRVGPTAVRDDGDRLKVSRGADQDGMQAALRMAMDRYGDRISVGGTPAFKEQIAQA
AAASKLPITFDDDALERRRQELLTQATTKENDHDRTDRGRAAGGGISGSGPAIAAIPGAARAGQPRAAAA
GRTTKPNVGRVGRKPPPQSQNRLRRLSQLGVVQLASGSEVLLPRHVPGDVEQQGAKPVDGLRRDIFGPGR
LSPQVAAAADKYIAEREGKRLKGFDIPKHSRYTDGQDGAATFAGIRQVEGQSLALLTRGEEVMVLPVDEP
TARRLKRLAVGDAITVTPQGSIKTTKGRSR
MIAKHVSMKTVQKSDFAGLVKYITDEQAKNERVGYVAVTNCHTDRPEVAITEVLNTQAQNTRSGADKTYH
LIVSFRPGEQPDDATLKAIEARICEGLGFGEHQRVSAVHHDTDNLHIHIAINKIHPTRYTILEPFNAYHT
LGQLCEKLEREYGLERDNHQGNKLVSEGRAQDMERHAGVESLQGWVKRECAADMQAAQSWEALHQVMRDN
GLELRERGNGFVIQAADGLAVKASSIGREFSKAKLEERLGTFEPSAEHLERAAQKPRKQYDKKPVRSRVN
TVELHAKYKADQLTISGSRTAEWEKARAKKDRLIEAAKRSGRLKRSTIKLISGAGVGKKLMYSATSKTMK
SEIEKINKQYLKERQEIHEKYQRRAWADWLRAKAAEGDQEALGALRARDAAQGLKGNTVAGTGPLRPQAA
AAEQDSVTKKGTIIYRVGPTAVRDDGDRLKVSRGADQDGMQAALRMAMDRYGDRISVGGTPAFKEQIAQA
AAASKLPITFDDDALERRRQELLTQATTKENDHDRTDRGRAAGGGISGSGPAIAAIPGAARAGQPRAAAA
GRTTKPNVGRVGRKPPPQSQNRLRRLSQLGVVQLASGSEVLLPRHVPGDVEQQGAKPVDGLRRDIFGPGR
LSPQVAAAADKYIAEREGKRLKGFDIPKHSRYTDGQDGAATFAGIRQVEGQSLALLTRGEEVMVLPVDEP
TARRLKRLAVGDAITVTPQGSIKTTKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | K4IYE5 |
T4CP
| ID | 15842 | GenBank | WP_015063532 |
| Name | t4cp2_E5P2_RS37345_SRS16|5 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69689.93 Da Isoelectric Point: 9.0430
>WP_015063532.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Burkholderiales]
MTKDRFNNAVGPQVRAEKEGPGKVIPALAVLSLGGGLQAATQFFAHDFKYQAALGAHINHIYPPWGIVQW
AGKWYGQYPDAFMRAGSIGVTITGVGLISLVVAKMVLANSSKVNEYLHGSARWANVRDIQAAGLIPRARS
FWQTITGKDAAASSGVYVGAWIDKKGKQHYLRHNGPEHVLCYAPTRSGKGVGLVVPTLLSWGHSAVITDL
KGELWALTAGWRQKHAKNKVLRFEPATASGSVCWNPLDEIRLGTENEVGDVQNLATLIVDPDGKGLESHW
QKTSQALLVGVILHALYKAKAEGTPATLPGVDAMLADPNRDIGELWMEMTTYSHVDGQVHPVVGSAARDM
MDRPDEEAGSVLSTAKSYLALYRDPIVARNVSKSEFKIKDLMHHDNAVSLYIVTQPNDKARLRPLVRVMV
NMIVRLLADKMDFENGRPVAHYKHRMLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRE
TGYGHDESITSNCHVQNAYPPNRVETAEHLSKLTGQTTVAKEQITTSGRRTSALLGQVSRTIQEVQRPLL
TPDECLRMPGPVKTEGGDIAKAGDMVVYVAGYPAIYGKQPLYFQDPTFQARAAIPAPKNSDKLRNSFVAP
AGEGITI
MTKDRFNNAVGPQVRAEKEGPGKVIPALAVLSLGGGLQAATQFFAHDFKYQAALGAHINHIYPPWGIVQW
AGKWYGQYPDAFMRAGSIGVTITGVGLISLVVAKMVLANSSKVNEYLHGSARWANVRDIQAAGLIPRARS
FWQTITGKDAAASSGVYVGAWIDKKGKQHYLRHNGPEHVLCYAPTRSGKGVGLVVPTLLSWGHSAVITDL
KGELWALTAGWRQKHAKNKVLRFEPATASGSVCWNPLDEIRLGTENEVGDVQNLATLIVDPDGKGLESHW
QKTSQALLVGVILHALYKAKAEGTPATLPGVDAMLADPNRDIGELWMEMTTYSHVDGQVHPVVGSAARDM
MDRPDEEAGSVLSTAKSYLALYRDPIVARNVSKSEFKIKDLMHHDNAVSLYIVTQPNDKARLRPLVRVMV
NMIVRLLADKMDFENGRPVAHYKHRMLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRE
TGYGHDESITSNCHVQNAYPPNRVETAEHLSKLTGQTTVAKEQITTSGRRTSALLGQVSRTIQEVQRPLL
TPDECLRMPGPVKTEGGDIAKAGDMVVYVAGYPAIYGKQPLYFQDPTFQARAAIPAPKNSDKLRNSFVAP
AGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15843 | GenBank | WP_015063520 |
| Name | t4cp2_E5P2_RS37460_SRS16|5 |
UniProt ID | _ |
| Length | 845 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 845 a.a. Molecular weight: 93707.73 Da Isoelectric Point: 6.3402
>WP_015063520.1 MULTISPECIES: VirB4 family type IV secretion/conjugal transfer ATPase [Burkholderiales]
MIEAIAIGIAALGALLLLILFARIRAVDAELKLKKHRSKDAGLADLLNYAAMVDDGVIVGKNGSFIASWL
YKGDDNASSTDEQREMVSFRINQALAGLGNGWMLHVDAVRRPAPNYTETGLSHFPDPVSAAMDEERRRLF
EGLGTMYEGYFIVTMTYFPPLLAQSKFVELMFDDEAKTPGNKARTQGLIDYFKRECTSIESRLSSAVKLQ
RLRGNKVVNEDGSTVTHDDLLRWLQFCITGMDHPVLLPSNPMYLDAVIGGQELFGGVVPKIGRKFVQVVA
IEGFPLESTPGVLTALGELPVEYRWSSRFIFMDSHEAVKHLDKFRKKWKQKIRGFFDQVFNTNTGSVDQD
AMSMVNDAESAIAEVNSGLVAVGYYTSVVVLMDEDREKLETSARQVEKAINRLGFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSTIWTGLNKAPCPMYPPLSPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLALIAAQLRRYQGMSIYAFDKGMSMYPLAKATGGKHFTVAADDDKLAFCPLQFLES
KSDRAWAMEWIDTILALNNVETTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIRQYTVDGNM
GHLLDAEEDGLSLTDFTVFEIEELMNLGEKYALPTLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRA
KIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMGLNARQIE
ILATAIPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVAAIKSLEAKFGDAWVHEWLAGRNLKLSD
YGVAA
MIEAIAIGIAALGALLLLILFARIRAVDAELKLKKHRSKDAGLADLLNYAAMVDDGVIVGKNGSFIASWL
YKGDDNASSTDEQREMVSFRINQALAGLGNGWMLHVDAVRRPAPNYTETGLSHFPDPVSAAMDEERRRLF
EGLGTMYEGYFIVTMTYFPPLLAQSKFVELMFDDEAKTPGNKARTQGLIDYFKRECTSIESRLSSAVKLQ
RLRGNKVVNEDGSTVTHDDLLRWLQFCITGMDHPVLLPSNPMYLDAVIGGQELFGGVVPKIGRKFVQVVA
IEGFPLESTPGVLTALGELPVEYRWSSRFIFMDSHEAVKHLDKFRKKWKQKIRGFFDQVFNTNTGSVDQD
AMSMVNDAESAIAEVNSGLVAVGYYTSVVVLMDEDREKLETSARQVEKAINRLGFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSTIWTGLNKAPCPMYPPLSPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLALIAAQLRRYQGMSIYAFDKGMSMYPLAKATGGKHFTVAADDDKLAFCPLQFLES
KSDRAWAMEWIDTILALNNVETTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIRQYTVDGNM
GHLLDAEEDGLSLTDFTVFEIEELMNLGEKYALPTLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRA
KIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMGLNARQIE
ILATAIPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVAAIKSLEAKFGDAWVHEWLAGRNLKLSD
YGVAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 26130..36727
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| E5P2_RS38115 | 22212..22373 | - | 162 | Protein_19 | antitoxin VbhA family protein | - |
| E5P2_RS37385 (G3W92_RS37370) | 22340..22621 | + | 282 | Protein_20 | helix-turn-helix domain-containing protein | - |
| E5P2_RS37390 (G3W92_RS37375) | 22578..22985 | - | 408 | WP_031944013 | hypothetical protein | - |
| E5P2_RS37395 (G3W92_RS37380) | 22982..23467 | - | 486 | WP_015063510 | thermonuclease family protein | - |
| E5P2_RS37400 (G3W92_RS37385) | 23469..23894 | - | 426 | WP_053057113 | OmpA family protein | - |
| E5P2_RS37405 (G3W92_RS37390) | 23908..24606 | - | 699 | WP_269474032 | TraX family protein | - |
| E5P2_RS37410 (G3W92_RS37395) | 24609..24872 | - | 264 | WP_162572153 | conjugal transfer protein TrbO | - |
| E5P2_RS37415 (G3W92_RS37400) | 24898..25503 | - | 606 | WP_162572154 | transglycosylase SLT domain-containing protein | - |
| E5P2_RS37420 (G3W92_RS37405) | 25517..26110 | - | 594 | WP_015063527 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| E5P2_RS37425 (G3W92_RS37410) | 26130..27863 | - | 1734 | WP_174259956 | P-type conjugative transfer protein TrbL | virB6 |
| E5P2_RS37430 (G3W92_RS37415) | 27874..28113 | - | 240 | WP_031944010 | entry exclusion lipoprotein TrbK | - |
| E5P2_RS37435 (G3W92_RS37420) | 28124..28897 | - | 774 | WP_031944011 | P-type conjugative transfer protein TrbJ | virB5 |
| E5P2_RS37440 (G3W92_RS37425) | 28916..30310 | - | 1395 | WP_015063524 | TrbI/VirB10 family protein | virB10 |
| E5P2_RS37445 (G3W92_RS37430) | 30316..30783 | - | 468 | WP_015063523 | TrbH Mating pair formation protein | - |
| E5P2_RS37450 (G3W92_RS37435) | 30786..31691 | - | 906 | WP_015063522 | P-type conjugative transfer protein TrbG | virB9 |
| E5P2_RS37455 (G3W92_RS37440) | 31704..32480 | - | 777 | WP_015063521 | conjugal transfer protein TrbF | virB8 |
| E5P2_RS37460 (G3W92_RS37445) | 32477..35014 | - | 2538 | WP_015063520 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| E5P2_RS37465 (G3W92_RS37450) | 35011..35322 | - | 312 | WP_011171720 | conjugal transfer protein TrbD | virB3 |
| E5P2_RS37470 (G3W92_RS37455) | 35325..35753 | - | 429 | WP_011171719 | IncP-type conjugal transfer pilin TrbC | virB2 |
| E5P2_RS37475 (G3W92_RS37460) | 35765..36727 | - | 963 | WP_011171718 | P-type conjugative transfer ATPase TrbB | virB11 |
| E5P2_RS37480 (G3W92_RS37465) | 36980..37357 | - | 378 | WP_011171717 | transcriptional regulator | - |
| E5P2_RS37485 (G3W92_RS37470) | 37678..38547 | + | 870 | WP_015063508 | plasmid replication initiator TrfA | - |
| E5P2_RS37490 (G3W92_RS37475) | 38916..40022 | + | 1107 | WP_162572156 | NotI family restriction endonuclease | - |
| E5P2_RS37495 (G3W92_RS37480) | 40122..41150 | + | 1029 | WP_162572157 | DNA adenine methylase | - |
Host bacterium
| ID | 21927 | GenBank | NZ_LR594670 |
| Plasmid name | SRS16|5 | Incompatibility group | - |
| Plasmid size | 71057 bp | Coordinate of oriT [Strand] | 6794..6888 [-] |
| Host baterium | Variovorax sp. SRS16 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9, AcrIF24 |