Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121443 |
| Name | oriT_E7429|3 |
| Organism | Enterococcus faecium isolate E7429 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LR135299 (28421..28612 [+], 192 nt) |
| oriT length | 192 nt |
| IRs (inverted repeats) | 106..112, 122..128 (ATTTTTT..AAAAAAT) 107..113, 120..126 (TTTTTTG..CAAAAAA) 93..98, 105..110 (AAAATA..TATTTT) 65..71, 75..81 (AGTTGGC..GCCAACT) 41..49, 52..60 (CACCTTCCT..AGGAAGGTG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 192 nt
>oriT_E7429|3
CCAAGGAATTAATACAAAGAGCATAAGGGAAAACTGATAGCACCTTCCTAAAGGAAGGTGGCTAAGTTGGCTGTGCCAACTGGTTTTCTTTCAAAATAGCTTCATATTTTTTGCTATCACAAAAAAATCCATTTTCGACCTATTTTCGGTCATAATATAGTACCTACTTTTGGTCATAGTTTCGTCCGTAGT
CCAAGGAATTAATACAAAGAGCATAAGGGAAAACTGATAGCACCTTCCTAAAGGAAGGTGGCTAAGTTGGCTGTGCCAACTGGTTTTCTTTCAAAATAGCTTCATATTTTTTGCTATCACAAAAAAATCCATTTTCGACCTATTTTCGGTCATAATATAGTACCTACTTTTGGTCATAGTTTCGTCCGTAGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file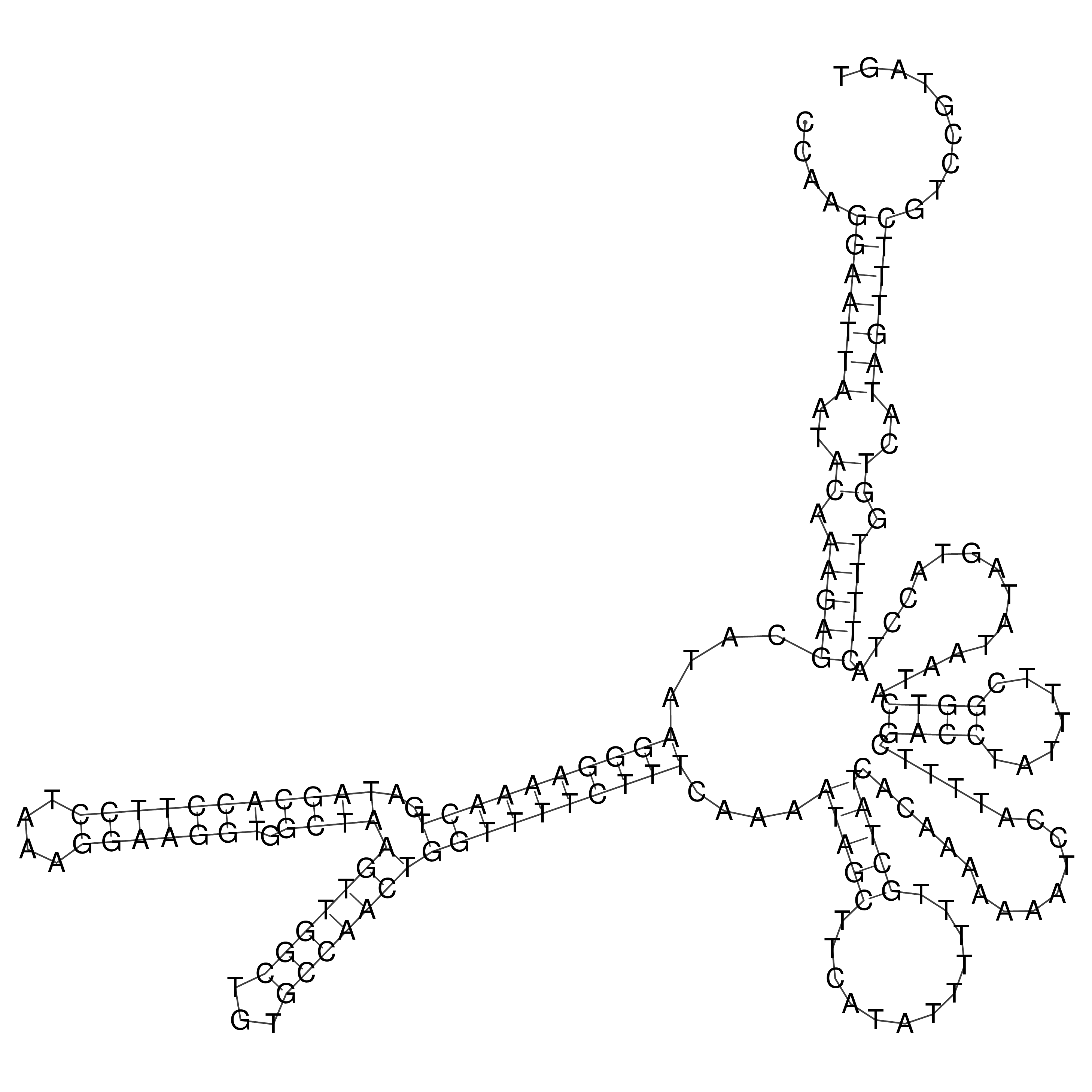
Relaxase
| ID | 13519 | GenBank | WP_002326870 |
| Name | mobP2_EQB73_RS16575_E7429|3 |
UniProt ID | B5U8V6 |
| Length | 506 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 506 a.a. Molecular weight: 60597.01 Da Isoelectric Point: 9.3485
>WP_002326870.1 MULTISPECIES: MobP2 family relaxase [Enterococcus]
MTEHIFNRKSASIILKAKFETGKSKKNNHMVELQKFVDYISRQEAIRQDKLDHDYSEDELNELARIEKAL
EKIEQETDQPVIKDLREMDKYIDYMTRKKAILENVDNEIVNGAFSNTKRYITKKDISKIKESVIEAKNNG
SVMFQDVISFDNDFLVREGYYNPETNELNENVLYEATKSMMGKMQEKEELVDPFWFATIHRNTEHIHIHV
TAMERKNTREIMEYDGVLQARGKRKQSTLDDMIFKFGSKILDRTNEFEKISKLRKEVPLELKQSVKDSLI
QLYVENNSRYDKELVKYLKELKKEIPSTTRGYNELPEETKEKIDEVTNYMTKDNPKRKEYDRMTKEIDEL
YQTTYGKRYNPQEYYLNRKKDFNARMGNAVVGQIKLLKQSEEKNLKSNELKNLLDGKKLNVKFDREKPEF
KTKYAKQSYDDFQKRLEERQLAREKNEVHFKKISEKFKDKKLIRKIDRVINDDLNMYRAKNDYEVAQRMI
EQEKMRQAREFEIGMN
MTEHIFNRKSASIILKAKFETGKSKKNNHMVELQKFVDYISRQEAIRQDKLDHDYSEDELNELARIEKAL
EKIEQETDQPVIKDLREMDKYIDYMTRKKAILENVDNEIVNGAFSNTKRYITKKDISKIKESVIEAKNNG
SVMFQDVISFDNDFLVREGYYNPETNELNENVLYEATKSMMGKMQEKEELVDPFWFATIHRNTEHIHIHV
TAMERKNTREIMEYDGVLQARGKRKQSTLDDMIFKFGSKILDRTNEFEKISKLRKEVPLELKQSVKDSLI
QLYVENNSRYDKELVKYLKELKKEIPSTTRGYNELPEETKEKIDEVTNYMTKDNPKRKEYDRMTKEIDEL
YQTTYGKRYNPQEYYLNRKKDFNARMGNAVVGQIKLLKQSEEKNLKSNELKNLLDGKKLNVKFDREKPEF
KTKYAKQSYDDFQKRLEERQLAREKNEVHFKKISEKFKDKKLIRKIDRVINDDLNMYRAKNDYEVAQRMI
EQEKMRQAREFEIGMN
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B5U8V6 |
T4CP
| ID | 15826 | GenBank | WP_060811444 |
| Name | t4cp2_EQB73_RS16640_E7429|3 |
UniProt ID | _ |
| Length | 952 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 952 a.a. Molecular weight: 108255.15 Da Isoelectric Point: 7.4751
>WP_060811444.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecium]
MKLVQNNEVKEIKNGFVKNSNKILGKLKKVGSFKKNKYHTGKIEGEDLPKSLQDKLNVPLVFIGVFLGSI
VFLAITYLLNFLKALLVAIGQSESGLLGVEFDPKFSLAYLKPLGDTKQYFMCAVVAVIGVAYLIYGKLNH
ESEKNLVYGQKGDSRLSTIKEIKRDYHEIPEKTEVYDGIGGVPISHYQDKYYIDRDTVNTVVLGASRSGK
GETFVVPLLDNLSRAKIQSNMVVNDPKGELFSASKETLEKRGYRVLVLNIDDPLESMSFNPLQLVIDSWA
NKDYHEASKRANTLTSMLFASGMSTDNEFFYKSAKSAVNAIILTIVEHCFNNDCIEKITMYNVAQMLNEL
GSLFYTDPKTGKEKNALDEYFNTLPQGNQAKIQYGSTSFAGDKAKGSILSTASQGIEMFTSDLFGKLTSK
MSIDLKEIGFPKSIQFKVNTNLVGKRISISFLRKENDVIRLIKKYRVKVKALGLCVLNFNEYLQDGDMLD
IRYEEEKSRAWYRIEFPKKQANSHDTSLVTKKFKSGKNYSDLEISTLRLKYTDQPTAVFMVIPDYDPSNH
VLASIFISQLYTELASNCKQTPDKKCFRRVHFLLDEFGNMPAIDNMDGIMTVCLGRNMLFDLVIQSYSQL
ETRYDKAFKTIKENCQNHILIMSNDEETIEEISKKCGHKTTINRSASSKHLDTDSSVTSSAEQERVITVE
RASQLIEGEQIILRNLHRQDTKRNKIRPFPIFNTKETNMPYRYQFLAEDFDTQRDINEIDIACEHINLSL
QDNQIPYARFIKDLKTRLEYSIINNIPISEEDYQDYLNLSGSQQTINEEELLKLVNTEVKSQDMEGQVLS
EQEMKINIYLKESQKEAIELVKVINGRLADENSADMVIKTINSQILDTPFIQSLINDSIAIHNVQESKDQ
IMILEQQLANAYELAVFKTKNSILKNKIIKLKNAMTKLAKAI
MKLVQNNEVKEIKNGFVKNSNKILGKLKKVGSFKKNKYHTGKIEGEDLPKSLQDKLNVPLVFIGVFLGSI
VFLAITYLLNFLKALLVAIGQSESGLLGVEFDPKFSLAYLKPLGDTKQYFMCAVVAVIGVAYLIYGKLNH
ESEKNLVYGQKGDSRLSTIKEIKRDYHEIPEKTEVYDGIGGVPISHYQDKYYIDRDTVNTVVLGASRSGK
GETFVVPLLDNLSRAKIQSNMVVNDPKGELFSASKETLEKRGYRVLVLNIDDPLESMSFNPLQLVIDSWA
NKDYHEASKRANTLTSMLFASGMSTDNEFFYKSAKSAVNAIILTIVEHCFNNDCIEKITMYNVAQMLNEL
GSLFYTDPKTGKEKNALDEYFNTLPQGNQAKIQYGSTSFAGDKAKGSILSTASQGIEMFTSDLFGKLTSK
MSIDLKEIGFPKSIQFKVNTNLVGKRISISFLRKENDVIRLIKKYRVKVKALGLCVLNFNEYLQDGDMLD
IRYEEEKSRAWYRIEFPKKQANSHDTSLVTKKFKSGKNYSDLEISTLRLKYTDQPTAVFMVIPDYDPSNH
VLASIFISQLYTELASNCKQTPDKKCFRRVHFLLDEFGNMPAIDNMDGIMTVCLGRNMLFDLVIQSYSQL
ETRYDKAFKTIKENCQNHILIMSNDEETIEEISKKCGHKTTINRSASSKHLDTDSSVTSSAEQERVITVE
RASQLIEGEQIILRNLHRQDTKRNKIRPFPIFNTKETNMPYRYQFLAEDFDTQRDINEIDIACEHINLSL
QDNQIPYARFIKDLKTRLEYSIINNIPISEEDYQDYLNLSGSQQTINEEELLKLVNTEVKSQDMEGQVLS
EQEMKINIYLKESQKEAIELVKVINGRLADENSADMVIKTINSQILDTPFIQSLINDSIAIHNVQESKDQ
IMILEQQLANAYELAVFKTKNSILKNKIIKLKNAMTKLAKAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 21870 | GenBank | NZ_LR135299 |
| Plasmid name | E7429|3 | Incompatibility group | - |
| Plasmid size | 58745 bp | Coordinate of oriT [Strand] | 28421..28612 [+] |
| Host baterium | Enterococcus faecium isolate E7429 |
Cargo genes
| Drug resistance gene | dfrG |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA1 |