Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121441 |
| Name | oriT_E7025|4 |
| Organism | Enterococcus faecium isolate E7025 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LR135229 (7891..7933 [+], 43 nt) |
| oriT length | 43 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 43 nt
>oriT_E7025|4
AATATCGCAACATGGGCCCATGTTGCTCTGCTCGCAAAAGAAA
AATATCGCAACATGGGCCCATGTTGCTCTGCTCGCAAAAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file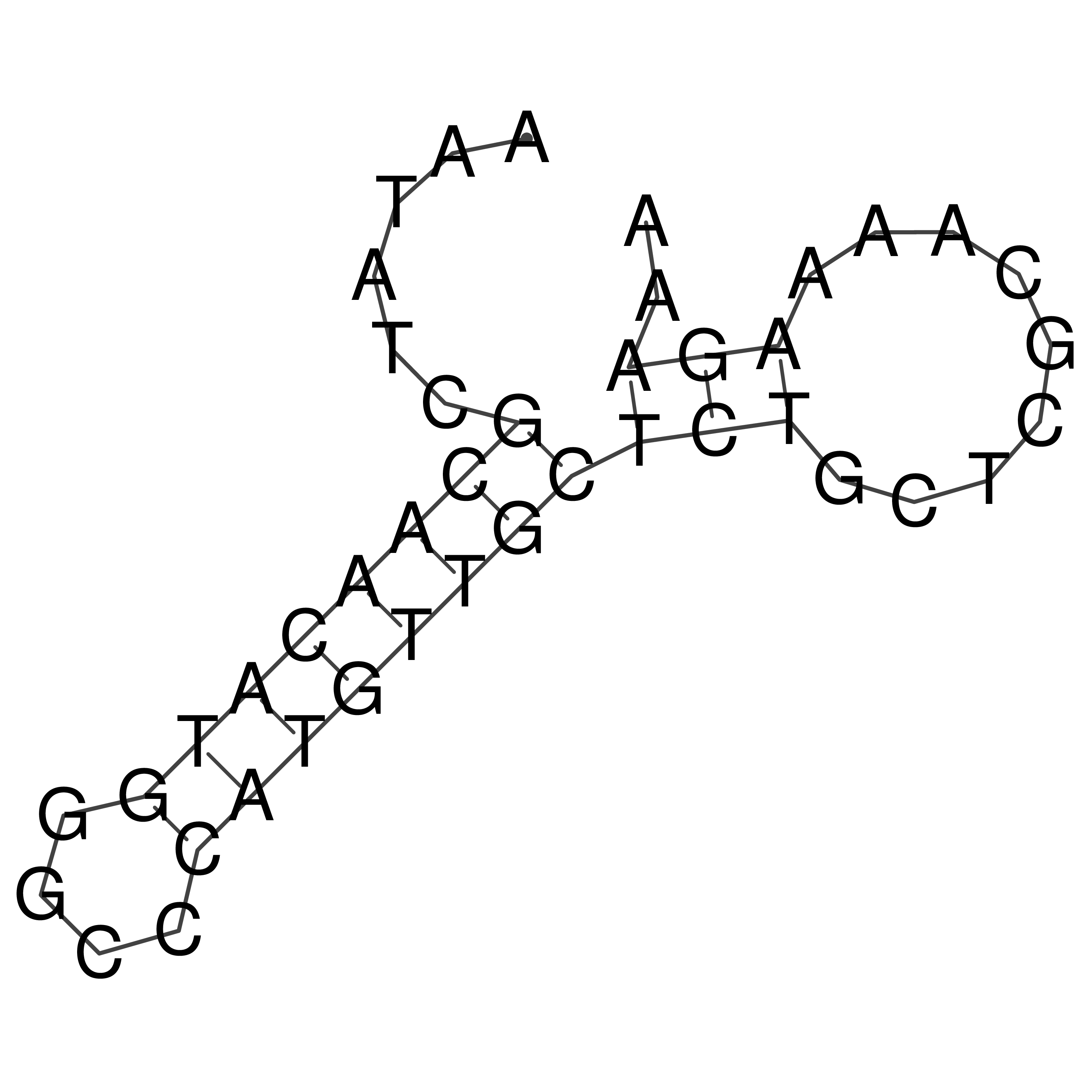
Relaxase
| ID | 13518 | GenBank | WP_127823964 |
| Name | Relaxase_EQB58_RS15740_E7025|4 |
UniProt ID | _ |
| Length | 555 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 555 a.a. Molecular weight: 63881.34 Da Isoelectric Point: 6.4731
>WP_127823964.1 relaxase/mobilization nuclease domain-containing protein [Enterococcus faecium]
MVYTKHFTIHSSKHLKQAEEYVENAAKTAIDSKRGIDSHLDNIFPYITSDKKIINKQLVSGYGIIDVYNS
AKEFLLTKQNAAFAKGKDLIFNPKTGKVVLDPQALEKNNAVLAHHIIQSFSPEDDLTPEQIHEIGRKTIL
EFTGGEYEFVIATHVDKAHIHNHIILNSTNTFTGNALRWQKGTKKAFEQVSDKIASKYGAKIIEKSPKNS
HKKYTMWQTENLFKNKIKSRLDFLLDHSSSIDDFKIKAEALDLQVDFSGKWAKYCLLDEPQIKNTRSRSL
SKKDPEKYNLDRIVERLKENSHEFSVQEVLERYEEKVDAVNNDFDYQLILESWQISHKTPKGYYVNVDFG
AENHGQIFIGAYKVDQLEDGNYSLFVKKNDYFYFMNDKSSDRNRYMTGYTLAKQLSLYNGTVPLKKEPVM
TTITELVDAINFLAEHGVTESKQMSRLENKLIESFELAEESLQKLDDKILQLNQLGKLIMEEKLEDTNQE
FPVLKEIGNKDELTFEDIQNELSSAKLSRKLLHEKMESTVEKINELHAIQTVAEQKSSEENLGKL
MVYTKHFTIHSSKHLKQAEEYVENAAKTAIDSKRGIDSHLDNIFPYITSDKKIINKQLVSGYGIIDVYNS
AKEFLLTKQNAAFAKGKDLIFNPKTGKVVLDPQALEKNNAVLAHHIIQSFSPEDDLTPEQIHEIGRKTIL
EFTGGEYEFVIATHVDKAHIHNHIILNSTNTFTGNALRWQKGTKKAFEQVSDKIASKYGAKIIEKSPKNS
HKKYTMWQTENLFKNKIKSRLDFLLDHSSSIDDFKIKAEALDLQVDFSGKWAKYCLLDEPQIKNTRSRSL
SKKDPEKYNLDRIVERLKENSHEFSVQEVLERYEEKVDAVNNDFDYQLILESWQISHKTPKGYYVNVDFG
AENHGQIFIGAYKVDQLEDGNYSLFVKKNDYFYFMNDKSSDRNRYMTGYTLAKQLSLYNGTVPLKKEPVM
TTITELVDAINFLAEHGVTESKQMSRLENKLIESFELAEESLQKLDDKILQLNQLGKLIMEEKLEDTNQE
FPVLKEIGNKDELTFEDIQNELSSAKLSRKLLHEKMESTVEKINELHAIQTVAEQKSSEENLGKL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15825 | GenBank | WP_127823995 |
| Name | t4cp1_EQB58_RS15960_E7025|4 |
UniProt ID | _ |
| Length | 392 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 392 a.a. Molecular weight: 45578.26 Da Isoelectric Point: 4.9830
>WP_127823995.1 VirD4-like conjugal transfer protein, CD1115 family, partial [Enterococcus faecium]
MQIYKKNIKNYLILSVLLFLIFYIIGNFFWLLPGSDYTAKMYFLFTEELNRYIEMNWLNFVLTPSLMAFC
FGILGFLCGMLAYVRDNDRGVYRYGEEHGSARYATVEEMSRFADQDPEKNIILTKNACMGISNRRLDLKY
QKNKNDVVIGGPGSGKTYTFIKPNIMQMNASFIVTDPKGLLVHETGSMLEKNGYKIKIFDLLNLSNSDSF
NVFNYIHTELDVDRVLEFITEGTKKSEQTGEDFWIQAEALLIRSFIAFLWFDGKDNDYMPNLSMVGDMLR
HTERKDPKVASPVENWFEEQNELRPNNYAYKQWKLFNDTYKAETRASVLAMAAGRYSVFDHDQVVDMVRE
DTMEIEKWNEEKTAVFISIPETSSAYNFLAAIFMATIMETLR
MQIYKKNIKNYLILSVLLFLIFYIIGNFFWLLPGSDYTAKMYFLFTEELNRYIEMNWLNFVLTPSLMAFC
FGILGFLCGMLAYVRDNDRGVYRYGEEHGSARYATVEEMSRFADQDPEKNIILTKNACMGISNRRLDLKY
QKNKNDVVIGGPGSGKTYTFIKPNIMQMNASFIVTDPKGLLVHETGSMLEKNGYKIKIFDLLNLSNSDSF
NVFNYIHTELDVDRVLEFITEGTKKSEQTGEDFWIQAEALLIRSFIAFLWFDGKDNDYMPNLSMVGDMLR
HTERKDPKVASPVENWFEEQNELRPNNYAYKQWKLFNDTYKAETRASVLAMAAGRYSVFDHDQVVDMVRE
DTMEIEKWNEEKTAVFISIPETSSAYNFLAAIFMATIMETLR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 39702..48848
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EQB58_RS15895 | 36201..38258 | - | 2058 | WP_127823987 | CAP domain-containing protein | - |
| EQB58_RS15900 | 38400..38606 | - | 207 | WP_086328743 | hypothetical protein | - |
| EQB58_RS15905 | 38782..39228 | + | 447 | WP_127824145 | hypothetical protein | - |
| EQB58_RS15910 | 39251..39676 | + | 426 | WP_010731367 | hypothetical protein | - |
| EQB58_RS15915 | 39702..39929 | + | 228 | WP_010731368 | hypothetical protein | prgF |
| EQB58_RS15920 | 39940..40737 | + | 798 | WP_127823989 | type IV secretion system protein | prgHb |
| EQB58_RS15925 | 40737..41087 | + | 351 | WP_127823990 | PrgI family mobile element protein | prgIc |
| EQB58_RS15930 | 41041..43416 | + | 2376 | WP_127823991 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| EQB58_RS15935 | 43428..46088 | + | 2661 | WP_127823992 | phage tail tip lysozyme | prgK |
| EQB58_RS15940 | 46106..46831 | + | 726 | WP_002332637 | hypothetical protein | prgL |
| EQB58_RS15945 | 46815..47060 | + | 246 | WP_127823993 | hypothetical protein | - |
| EQB58_RS15950 | 47084..48340 | + | 1257 | WP_127823994 | hypothetical protein | - |
| EQB58_RS15955 | 48363..48848 | + | 486 | WP_002332616 | DUF3801 domain-containing protein | gbs1365 |
| EQB58_RS15960 | 48848..50023 | + | 1176 | WP_127823995 | VirD4-like conjugal transfer protein, CD1115 family | - |
Host bacterium
| ID | 21868 | GenBank | NZ_LR135229 |
| Plasmid name | E7025|4 | Incompatibility group | - |
| Plasmid size | 50023 bp | Coordinate of oriT [Strand] | 7891..7933 [+] |
| Host baterium | Enterococcus faecium isolate E7025 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA1 |