Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121344 |
| Name | oriT_18ISCm|unnamed3 |
| Organism | Carnobacterium maltaromaticum strain 18ISCm |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP045043 (60359..60394 [-], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_18ISCm|unnamed3
CACACGTATGTGGGTACAGTTTCCCTTATGCTCTTT
CACACGTATGTGGGTACAGTTTCCCTTATGCTCTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file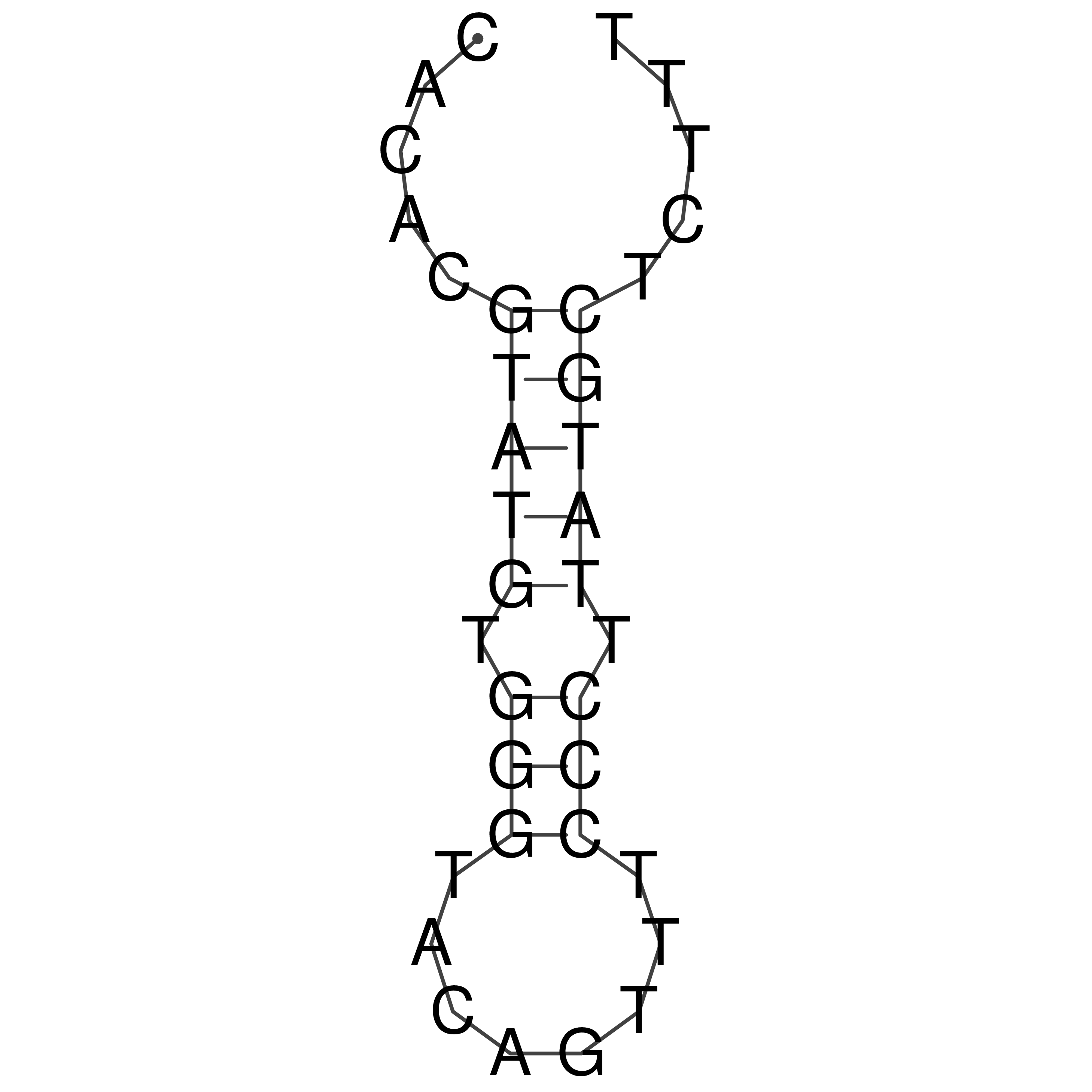
Relaxase
| ID | 13468 | GenBank | WP_157456903 |
| Name | mobP2_F9281_RS18780_18ISCm|unnamed3 |
UniProt ID | _ |
| Length | 413 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 413 a.a. Molecular weight: 48024.54 Da Isoelectric Point: 9.4699
>WP_157456903.1 MobP2 family relaxase [Carnobacterium maltaromaticum]
MAKGKPAIVMVAKFVVHSDAKFNSYVNYMDRTEAQRSEKFNEFNAFTFDGYHEYMENPYKSTGLFSKDYS
VMGDREKQLLKDKFTVAQENKSPMWQTVFSFDNAWLEEQGLMDGKVHYLNEDPIKEAVTLAMNEQMRLMK
MGDTANWTAAVHYNTDNVHVHVALVEEVPTRQKFIYGGEEQYKAKIPPKVMDRMKSTFSNSMVNRDMELA
RISYLIRTELKLGTGKDGLRDVRVRKTLTNLLNELPEDKRLWQYGNTAMIPFRNELNYVTDLILKDENPH
ALKELDGRLEDEMNFFKRLYGEGTKEFERFKEYKSNKLAELYKDNGNSILKELKVIQRNGGIEKVGQGEK
RRIQSSGKSFVSKKNVSAIKKAFGKNKQDYLGKLAYQRLQAEKENAAYQQKQNQETDRGYGYD
MAKGKPAIVMVAKFVVHSDAKFNSYVNYMDRTEAQRSEKFNEFNAFTFDGYHEYMENPYKSTGLFSKDYS
VMGDREKQLLKDKFTVAQENKSPMWQTVFSFDNAWLEEQGLMDGKVHYLNEDPIKEAVTLAMNEQMRLMK
MGDTANWTAAVHYNTDNVHVHVALVEEVPTRQKFIYGGEEQYKAKIPPKVMDRMKSTFSNSMVNRDMELA
RISYLIRTELKLGTGKDGLRDVRVRKTLTNLLNELPEDKRLWQYGNTAMIPFRNELNYVTDLILKDENPH
ALKELDGRLEDEMNFFKRLYGEGTKEFERFKEYKSNKLAELYKDNGNSILKELKVIQRNGGIEKVGQGEK
RRIQSSGKSFVSKKNVSAIKKAFGKNKQDYLGKLAYQRLQAEKENAAYQQKQNQETDRGYGYD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15776 | GenBank | WP_157456915 |
| Name | t4cp2_F9281_RS18840_18ISCm|unnamed3 |
UniProt ID | _ |
| Length | 853 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 853 a.a. Molecular weight: 97224.62 Da Isoelectric Point: 4.8916
>WP_157456915.1 VirD4-like conjugal transfer protein, CD1115 family [Carnobacterium maltaromaticum]
MVIDKVSEIFQKFDQKKAQWIVATALFVIATLLGNLMSSGLKTLDNLFQLNPQTKTVSDFIGFHMIDFFP
LYVILYGIAGASFFVVWTQLKASYESMEQGNKGTARWTTLTELYEQYTQVPKLAKSDDETYPGLSGTLIA
QDGDVSFIDEGPAHNKITGRSRSGKDQTKVLPDLDLYSRAEEKPHIISVSTKYEVFAAAKERLEKRGYEV
SLFNLIHLERSLMFNPLELIKKAYMKGDVDEAIELCKTFSFPLYHNEEAKEPVWEETAMALVNAIILALC
YEFIEKSENPKLTEKYVTLYSAVMMLIELGATDKHGNTLLDKYFASLEPDNPARMEYSTVQFADGQMRSS
IFASTQAKLRNFIAPKVAKMMSKSNFGFDRFLGFEGAYTTLIVDEMADSEVDYRENPFKAFDLGSFKTGS
EVPLPIVEPVSSQSKMISGFAVPGSTVTVILPTLENATGEFEQLSVTATKSGTFSIKLEKQIESNQAVKV
NMAIIKPQAVFMVLPDYIETNYIIASTWIQQVYFVTSQYASENGDKLKKRIRAQLNEFGNLPAFSNIPSI
ISVGAGRGFLVDFYLQNDNQLRLKYDETHAKIIESEAMNTFFIFSKDKLTRDNFSEQLGETEYISKSRSG
GLFSFHKQYTETVETRPLMKSDELGRLMEGETVVDRSVKRRDLKGNKIVPHPIWNRGKTSYSYAYEYLTE
QFPNDRNWRQLDMPKVTDIPLKKYGEEFVKRIRRPLNKLLKEESEQENRVELDDIMSHEDAVLDKEYKGF
GTGESAAEFEDIQEVKLHVERKLMSEAFSSEDIYVMYQAAIGLYPEKTGEIDSMDYVEEYQDFLALPEHR
ELNEKYAKLGFFN
MVIDKVSEIFQKFDQKKAQWIVATALFVIATLLGNLMSSGLKTLDNLFQLNPQTKTVSDFIGFHMIDFFP
LYVILYGIAGASFFVVWTQLKASYESMEQGNKGTARWTTLTELYEQYTQVPKLAKSDDETYPGLSGTLIA
QDGDVSFIDEGPAHNKITGRSRSGKDQTKVLPDLDLYSRAEEKPHIISVSTKYEVFAAAKERLEKRGYEV
SLFNLIHLERSLMFNPLELIKKAYMKGDVDEAIELCKTFSFPLYHNEEAKEPVWEETAMALVNAIILALC
YEFIEKSENPKLTEKYVTLYSAVMMLIELGATDKHGNTLLDKYFASLEPDNPARMEYSTVQFADGQMRSS
IFASTQAKLRNFIAPKVAKMMSKSNFGFDRFLGFEGAYTTLIVDEMADSEVDYRENPFKAFDLGSFKTGS
EVPLPIVEPVSSQSKMISGFAVPGSTVTVILPTLENATGEFEQLSVTATKSGTFSIKLEKQIESNQAVKV
NMAIIKPQAVFMVLPDYIETNYIIASTWIQQVYFVTSQYASENGDKLKKRIRAQLNEFGNLPAFSNIPSI
ISVGAGRGFLVDFYLQNDNQLRLKYDETHAKIIESEAMNTFFIFSKDKLTRDNFSEQLGETEYISKSRSG
GLFSFHKQYTETVETRPLMKSDELGRLMEGETVVDRSVKRRDLKGNKIVPHPIWNRGKTSYSYAYEYLTE
QFPNDRNWRQLDMPKVTDIPLKKYGEEFVKRIRRPLNKLLKEESEQENRVELDDIMSHEDAVLDKEYKGF
GTGESAAEFEDIQEVKLHVERKLMSEAFSSEDIYVMYQAAIGLYPEKTGEIDSMDYVEEYQDFLALPEHR
ELNEKYAKLGFFN
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 21771 | GenBank | NZ_CP045043 |
| Plasmid name | 18ISCm|unnamed3 | Incompatibility group | - |
| Plasmid size | 107821 bp | Coordinate of oriT [Strand] | 60359..60394 [-] |
| Host baterium | Carnobacterium maltaromaticum strain 18ISCm |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA1 |