Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121324 |
| Name | oriT_p12084-FII |
| Organism | Klebsiella michiganensis strain 12084 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MZ398018 (153646..153695 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_p12084-FII
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file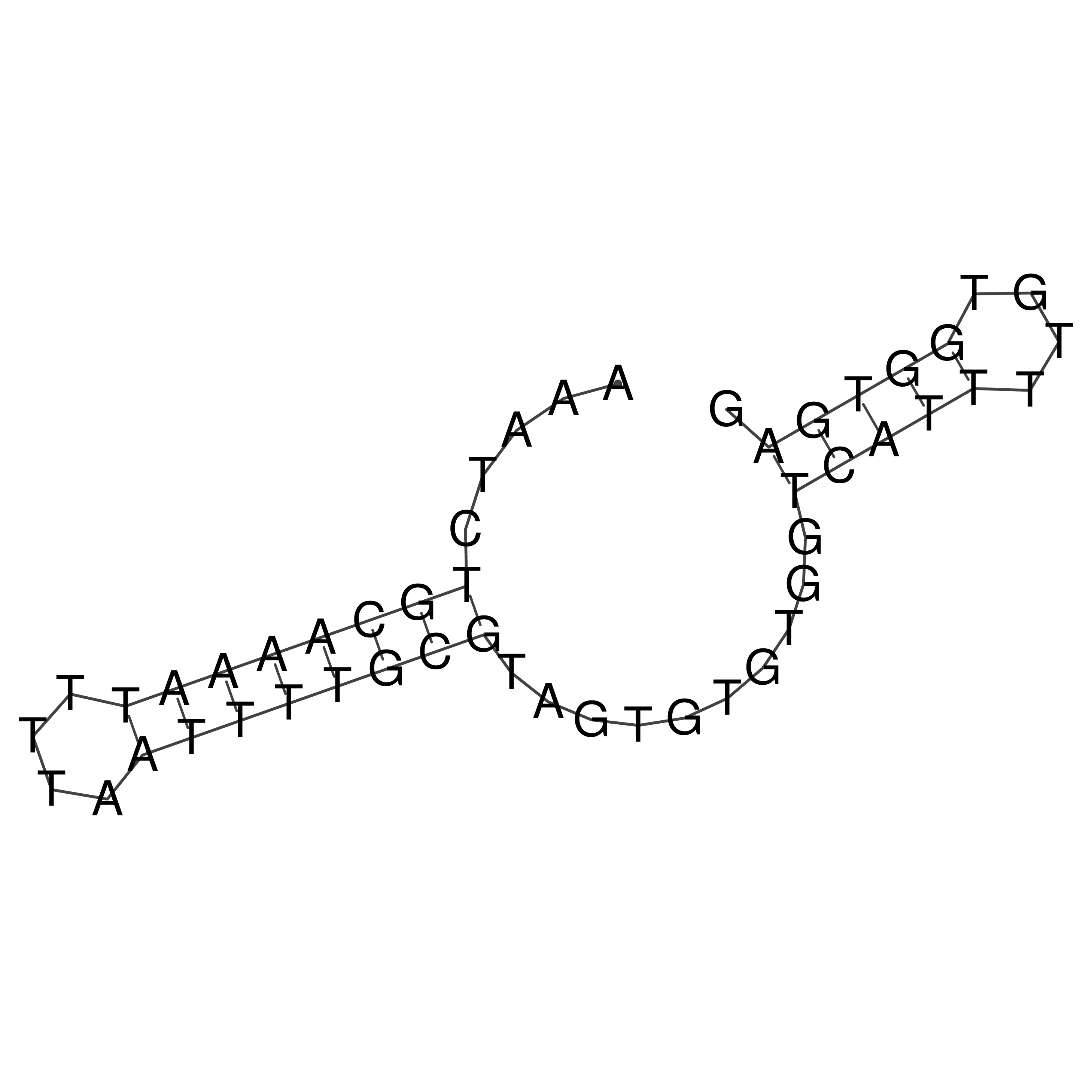
Relaxase
| ID | 13460 | GenBank | WP_040224080 |
| Name | traI_LO867_RS01005_p12084-FII |
UniProt ID | A0A7W3BA55 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190352.82 Da Isoelectric Point: 5.9133
>WP_040224080.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVIGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMNVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASETLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVIGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMNVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASETLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15763 | GenBank | WP_023329042 |
| Name | traD_LO867_RS01000_p12084-FII |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85943.90 Da Isoelectric Point: 5.0577
>WP_023329042.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRVDARLSALLEARESEGSLARTLFMPDAPASGPADTNSHAGEQPEPISQPA
PADMTVSPAPVKAPPTTKSPAAEPSVRATEPPVLRVTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRVDARLSALLEARESEGSLARTLFMPDAPASGPADTNSHAGEQPEPISQPA
PADMTVSPAPVKAPPTTKSPAAEPSVRATEPPVLRVTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 153088..182332
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LO867_RS00775 | 148106..148456 | + | 351 | WP_004182068 | hypothetical protein | - |
| LO867_RS00780 | 149092..149448 | + | 357 | WP_004152717 | hypothetical protein | - |
| LO867_RS00785 | 149509..149721 | + | 213 | WP_004182070 | hypothetical protein | - |
| LO867_RS00790 | 149732..149956 | + | 225 | WP_020325128 | hypothetical protein | - |
| LO867_RS00795 | 150037..150357 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| LO867_RS00800 | 150347..150625 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| LO867_RS00805 | 150626..151039 | + | 414 | WP_004182074 | type II toxin-antitoxin system HigA family antitoxin | - |
| LO867_RS00810 | 151872..152693 | + | 822 | WP_004182076 | DUF932 domain-containing protein | - |
| LO867_RS00815 | 152726..153055 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| LO867_RS00820 | 153088..153573 | - | 486 | WP_004178063 | transglycosylase SLT domain-containing protein | virB1 |
| LO867_RS00825 | 153964..154380 | + | 417 | WP_072145360 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LO867_RS00830 | 154580..155299 | + | 720 | WP_016831034 | conjugal transfer protein TrbJ | - |
| LO867_RS00835 | 155432..155638 | + | 207 | WP_171773970 | TraY domain-containing protein | - |
| LO867_RS00840 | 155700..156068 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| LO867_RS00845 | 156082..156387 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| LO867_RS00850 | 156407..156973 | + | 567 | WP_016831035 | type IV conjugative transfer system protein TraE | traE |
| LO867_RS00855 | 156960..157700 | + | 741 | WP_014386193 | type-F conjugative transfer system secretin TraK | traK |
| LO867_RS00860 | 157700..159124 | + | 1425 | WP_023158013 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LO867_RS00865 | 159117..159713 | + | 597 | Protein_175 | conjugal transfer pilus-stabilizing protein TraP | - |
| LO867_RS00870 | 159736..160320 | + | 585 | WP_014386196 | type IV conjugative transfer system lipoprotein TraV | traV |
| LO867_RS00875 | 160452..160862 | + | 411 | WP_016831039 | hypothetical protein | - |
| LO867_RS00880 | 160867..161156 | + | 290 | Protein_178 | hypothetical protein | - |
| LO867_RS00885 | 161180..161398 | + | 219 | WP_014386199 | hypothetical protein | - |
| LO867_RS00890 | 161399..161710 | + | 312 | WP_014386200 | hypothetical protein | - |
| LO867_RS00895 | 161777..162181 | + | 405 | WP_016831042 | hypothetical protein | - |
| LO867_RS00900 | 162224..162613 | + | 390 | WP_023158009 | hypothetical protein | - |
| LO867_RS00905 | 162621..163019 | + | 399 | WP_023158008 | hypothetical protein | - |
| LO867_RS00910 | 163091..165730 | + | 2640 | WP_023158007 | type IV secretion system protein TraC | virb4 |
| LO867_RS00915 | 165730..166119 | + | 390 | WP_004167468 | type-F conjugative transfer system protein TrbI | - |
| LO867_RS00920 | 166119..166754 | + | 636 | WP_023158006 | type-F conjugative transfer system protein TraW | traW |
| LO867_RS00925 | 166789..167190 | + | 402 | WP_023158005 | hypothetical protein | - |
| LO867_RS00930 | 167187..168176 | + | 990 | WP_015632509 | conjugal transfer pilus assembly protein TraU | traU |
| LO867_RS00935 | 168452..169099 | + | 648 | WP_015632510 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LO867_RS00940 | 169158..171113 | + | 1956 | WP_015065533 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LO867_RS00945 | 171145..171399 | + | 255 | WP_015065558 | conjugal transfer protein TrbE | - |
| LO867_RS00950 | 171377..171625 | + | 249 | WP_004152675 | hypothetical protein | - |
| LO867_RS00955 | 171638..171964 | + | 327 | WP_015065534 | hypothetical protein | - |
| LO867_RS00960 | 171985..172737 | + | 753 | WP_015065535 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LO867_RS00965 | 172748..172987 | + | 240 | WP_023158003 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LO867_RS00970 | 172959..173516 | + | 558 | WP_023158002 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LO867_RS00975 | 173562..174005 | + | 444 | WP_023158001 | F-type conjugal transfer protein TrbF | - |
| LO867_RS00980 | 173983..175362 | + | 1380 | WP_023158000 | conjugal transfer pilus assembly protein TraH | traH |
| LO867_RS00985 | 175362..178211 | + | 2850 | WP_023329044 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LO867_RS00990 | 178217..178744 | + | 528 | WP_029498351 | conjugal transfer protein TraS | - |
| LO867_RS00995 | 178934..179665 | + | 732 | WP_004152629 | conjugal transfer complement resistance protein TraT | - |
| LO867_RS01000 | 180023..182332 | + | 2310 | WP_023329042 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 21751 | GenBank | NZ_MZ398018 |
| Plasmid name | p12084-FII | Incompatibility group | IncFII |
| Plasmid size | 193645 bp | Coordinate of oriT [Strand] | 153646..153695 [-] |
| Host baterium | Klebsiella michiganensis strain 12084 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merF, merP, merT, merR2, arsC, arsB, arsR, arsD, arsA, arsH, pcoE, pcoR, pcoD, pcoC, pcoB, pcoA, silP, silA, silB, silF, silC, silR, silS, silE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |