Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121304 |
| Name | oriT_pPS00262_4A.4 |
| Organism | Klebsiella variicola strain PS00262.4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP127500 (27760..27808 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pPS00262_4A.4
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file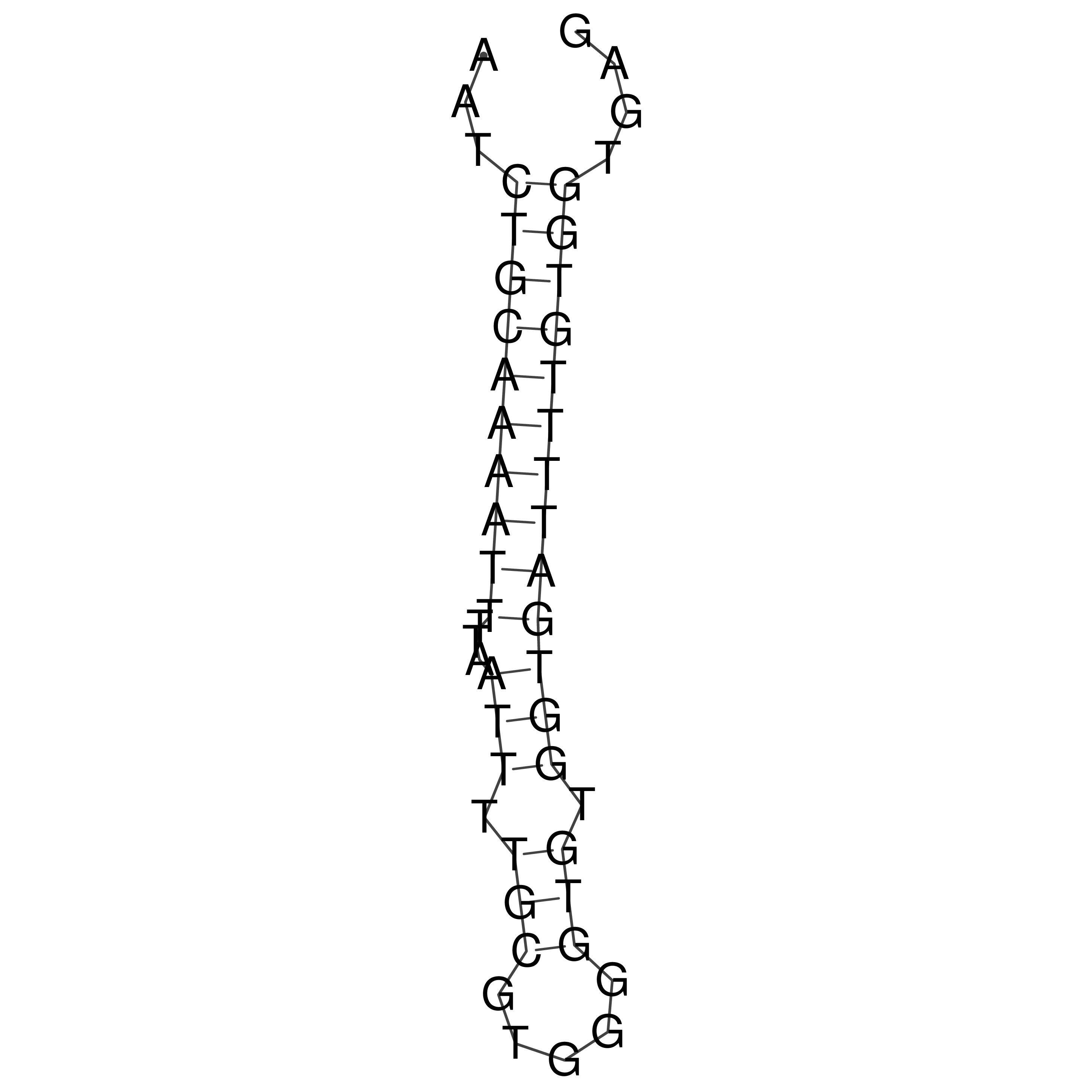
Relaxase
| ID | 13444 | GenBank | WP_110219713 |
| Name | traI_QQ383_RS31725_pPS00262_4A.4 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190646.70 Da Isoelectric Point: 6.2582
>WP_110219713.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MMSIGSVKSADKAADYYTNKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGRLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGLLTVQRPGQKTTQTLTVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAVGGGRAVPSGDTDQLPPIASGQPFRLTQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
NALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASERYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSADKAADYYTNKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGRLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGLLTVQRPGQKTTQTLTVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAVGGGRAVPSGDTDQLPPIASGQPFRLTQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
NALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASERYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7417 | GenBank | WP_020805752 |
| Name | WP_020805752_pPS00262_4A.4 |
UniProt ID | A0A377TIM3 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 14772.81 Da Isoelectric Point: 4.5715
>WP_020805752.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Gammaproteobacteria]
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A377TIM3 |
T4CP
| ID | 15743 | GenBank | WP_110219714 |
| Name | traD_QQ383_RS31720_pPS00262_4A.4 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 85407.28 Da Isoelectric Point: 5.0101
>WP_110219714.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQHTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGERPELASQPA
PAEVTVSPAPVKAPATTTMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAASTASSAGNPAAAAGGTEQEL
AQQSVEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINRSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQHTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGERPELASQPA
PAEVTVSPAPVKAPATTTMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAASTASSAGNPAAAAGGTEQEL
AQQSVEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINRSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 30187..55809
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QQ383_RS31535 (QQ383_31505) | 26640..27170 | + | 531 | WP_065954381 | antirestriction protein | - |
| QQ383_RS31540 (QQ383_31510) | 27208..27645 | - | 438 | WP_065954401 | transglycosylase SLT domain-containing protein | - |
| QQ383_RS31545 (QQ383_31515) | 28119..28511 | + | 393 | WP_020805752 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| QQ383_RS31550 (QQ383_31520) | 28749..29450 | + | 702 | WP_224324939 | hypothetical protein | - |
| QQ383_RS31555 (QQ383_31525) | 29536..29736 | + | 201 | WP_048289787 | TraY domain-containing protein | - |
| QQ383_RS31560 (QQ383_31530) | 29805..30173 | + | 369 | WP_020316649 | type IV conjugative transfer system pilin TraA | - |
| QQ383_RS31565 (QQ383_31535) | 30187..30492 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| QQ383_RS31570 (QQ383_31540) | 30512..31078 | + | 567 | WP_020316627 | type IV conjugative transfer system protein TraE | traE |
| QQ383_RS31575 (QQ383_31545) | 31065..31805 | + | 741 | WP_020804840 | type-F conjugative transfer system secretin TraK | traK |
| QQ383_RS31580 (QQ383_31550) | 31805..33229 | + | 1425 | WP_023316397 | F-type conjugal transfer pilus assembly protein TraB | traB |
| QQ383_RS31585 (QQ383_31555) | 33222..33818 | + | 597 | WP_023316398 | conjugal transfer pilus-stabilizing protein TraP | - |
| QQ383_RS31590 (QQ383_31560) | 33841..34410 | + | 570 | WP_086893255 | type IV conjugative transfer system lipoprotein TraV | traV |
| QQ383_RS31595 (QQ383_31565) | 34542..34952 | + | 411 | WP_020805613 | hypothetical protein | - |
| QQ383_RS31600 (QQ383_31570) | 34957..35241 | + | 285 | WP_119173789 | hypothetical protein | - |
| QQ383_RS31605 (QQ383_31575) | 35265..35483 | + | 219 | WP_124038229 | hypothetical protein | - |
| QQ383_RS31610 (QQ383_31580) | 35484..35795 | + | 312 | WP_124038230 | hypothetical protein | - |
| QQ383_RS31615 (QQ383_31585) | 35862..36266 | + | 405 | WP_023316404 | hypothetical protein | - |
| QQ383_RS31620 (QQ383_31590) | 36263..36553 | + | 291 | WP_020323516 | hypothetical protein | - |
| QQ383_RS31625 (QQ383_31595) | 36561..36959 | + | 399 | WP_072156904 | hypothetical protein | - |
| QQ383_RS31630 (QQ383_31600) | 37031..39670 | + | 2640 | WP_042946320 | type IV secretion system protein TraC | virb4 |
| QQ383_RS31635 (QQ383_31605) | 39670..40059 | + | 390 | WP_086893260 | type-F conjugative transfer system protein TrbI | - |
| QQ383_RS31640 (QQ383_31610) | 40059..40685 | + | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| QQ383_RS31645 (QQ383_31615) | 40729..41688 | + | 960 | WP_168712236 | conjugal transfer pilus assembly protein TraU | traU |
| QQ383_RS31650 (QQ383_31620) | 41701..42339 | + | 639 | WP_040107937 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| QQ383_RS31655 (QQ383_31625) | 42398..44353 | + | 1956 | WP_110219676 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| QQ383_RS31660 (QQ383_31630) | 44386..44667 | + | 282 | WP_022644736 | hypothetical protein | - |
| QQ383_RS31665 (QQ383_31635) | 44657..44884 | + | 228 | WP_012540032 | conjugal transfer protein TrbE | - |
| QQ383_RS31670 (QQ383_31640) | 44895..45221 | + | 327 | WP_020323521 | hypothetical protein | - |
| QQ383_RS31675 (QQ383_31645) | 45242..45994 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| QQ383_RS31680 (QQ383_31650) | 46005..46244 | + | 240 | WP_023340931 | type-F conjugative transfer system pilin chaperone TraQ | - |
| QQ383_RS31685 (QQ383_31655) | 46216..46773 | + | 558 | WP_086893263 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| QQ383_RS31690 (QQ383_31660) | 46819..47262 | + | 444 | WP_049248035 | F-type conjugal transfer protein TrbF | - |
| QQ383_RS31695 (QQ383_31665) | 47240..48619 | + | 1380 | WP_072198238 | conjugal transfer pilus assembly protein TraH | traH |
| QQ383_RS31700 (QQ383_31670) | 48619..51462 | + | 2844 | WP_110219715 | conjugal transfer mating-pair stabilization protein TraG | traG |
| QQ383_RS31705 (QQ383_31675) | 51473..52006 | + | 534 | WP_224324940 | hypothetical protein | - |
| QQ383_RS31710 (QQ383_31680) | 52417..53148 | + | 732 | WP_004152629 | conjugal transfer complement resistance protein TraT | - |
| QQ383_RS31715 (QQ383_31685) | 53339..53431 | + | 93 | Protein_75 | hypothetical protein | - |
| QQ383_RS31720 (QQ383_31690) | 53509..55809 | + | 2301 | WP_110219714 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 21732 | GenBank | NZ_CP127500 |
| Plasmid name | pPS00262_4A.4 | Incompatibility group | IncFIA |
| Plasmid size | 134238 bp | Coordinate of oriT [Strand] | 27760..27808 [-] |
| Host baterium | Klebsiella variicola strain PS00262.4 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merF, merP, merT, merR2, arsC, arsB |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |