Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121247 |
| Name | oriT_pWSM1455_1 |
| Organism | Rhizobium laguerreae strain WSM1455 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP088091 (744893..744921 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pWSM1455_1
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file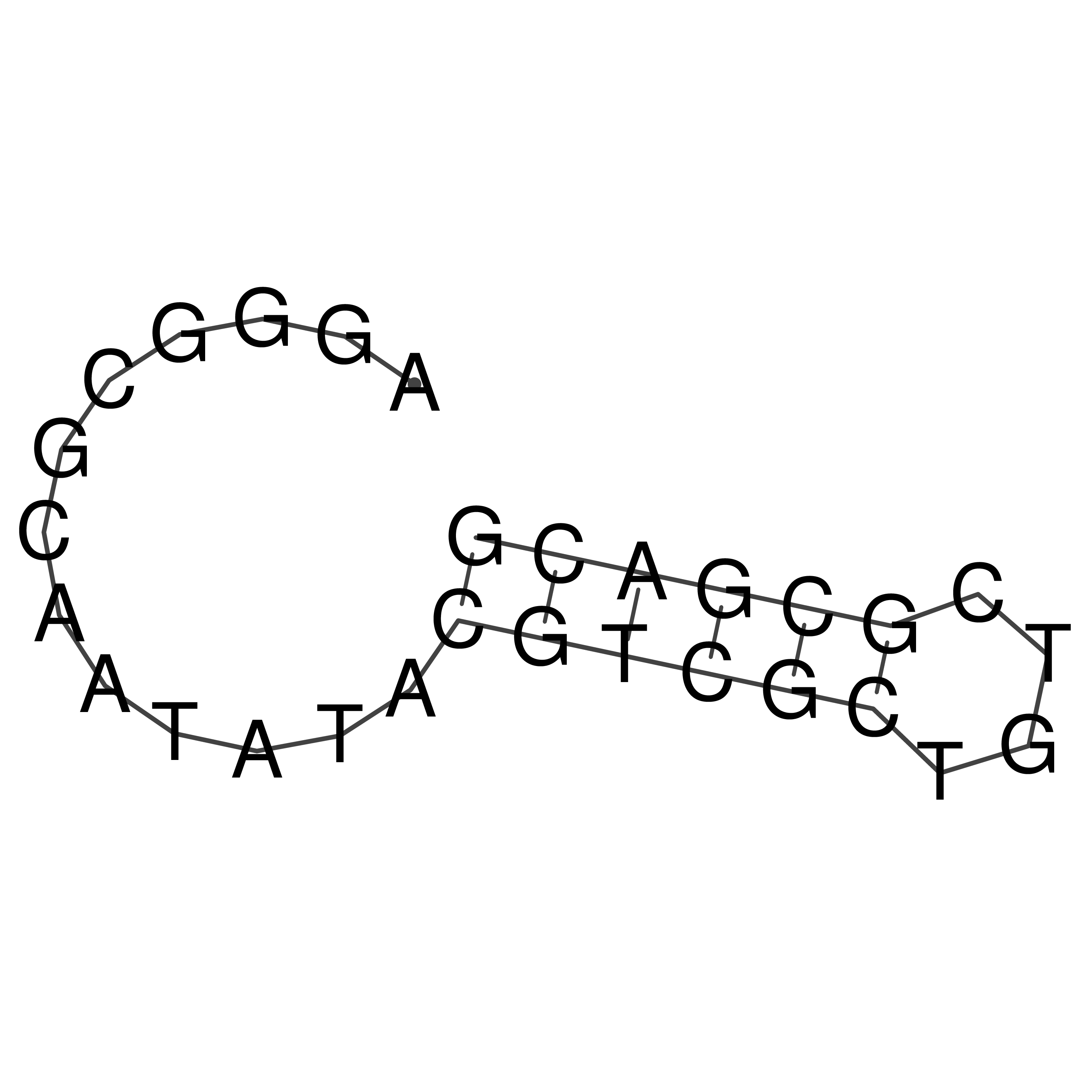
Relaxase
| ID | 13418 | GenBank | WP_231285546 |
| Name | traA_RlegWSM1455_RS27755_pWSM1455_1 |
UniProt ID | _ |
| Length | 1545 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1545 a.a. Molecular weight: 170547.81 Da Isoelectric Point: 6.7506
>WP_231285546.1 Ti-type conjugative transfer relaxase TraA [Rhizobium laguerreae]
MAIMFVRAQVISRGVGRSIVSAAAYRHRARMMDEQAGTSFSYRGGRAELVHEELALPTQIPEWLRTAIDG
RTVAAASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSEGMVADWVYHDKDG
NPHIHLMTALRPLTQEGFGRKKVAVTGEDGAPLRVVTPDRPKGRIVYKLWAGDKETMKAWKIAWAETANR
HLALAGHEIRIDGRSYAEQGLDGIAQRHLGPEKAALSRRGATMYFAPADLARRQEMADRLLAEPEFLLKR
LGNERSTFDERDIAKALHRYVDDPSDFANIRAGLMASDDLVILKPQEVDPETGKVAEPAVFTTRDILRVE
YDMARSAQVLSERRGFAVSAESVAAAIENVETADPEKSFRLDPEQVDAVRHVAGDNGIAAVVGLAGAGKS
TLLAAARVAWESGGRRVIGAALAGKAAEGLEDSSGISSRTLAAWELIWANGHETLHRGDVLVIDEAGMVS
SQQMARVLKLVEQARAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREEWAREASRLFARGE
IEKGLDAYAGHGHLVEAETRAETIDRIVADWTEARKQAIRNSTVVANDGRLRGDALLVLAHTNQDVKRLN
EALRSVMTGEGALGEGRSFRTERGAREFAAGDRMIFLENARFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GDTLLTVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVERTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEAKPEWGRKPHVDHAAGVTGELVETGQAKFRPNDEDADDSPYADVKTDDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVEKVKVQVPIVDEQTGQKRFEEREVDRNVWTAKRIETAAERQERIERESHRP
DVFKQLVERLSRSGAKTTTLDFESEAGYRAYADDFARRRGLDHLSLAAAGVEESLSRRWAGIAERREQLA
KLWERASMALGFTIERERRVAYSEERAEPTPVLEADVVSGRYLIPPATGFTRSVEEDARLAQLASPAGKE
REAILRPLLEKIYRDPDAALVALNALSSDASIEPRRLVDDLAATPDRLGRLRGSDLIVDGRAARDELNAA
ITALTELLPLARAHATEFRRNGERFDLGEQQRRTHMSLPIPALSEKAMARLTEIEAVRERGGDDAYKTAF
ALVTEDRSIVQEIKAVSEALTARFGWSAFTPNADAVAERNMVERMPEDLASIRGEKLTRLFEAVKRFADE
QHLAERRNRSKIVAAASADQGMGTDKENAAVLPMLAAVTDFKTPVDEEARSRVLSAPLYRQQRAALAEAA
RVIWRDPAGAVGKIEELLAKGFAGERIAAAVTNDPAAYGALRGSDRLMDRMLAAGRERKDAVQAMPEAAA
RLRALGSAYVNVLDGERRAIAEERRRMTVAIPGLSKAAEEVLMRLTAEAKNNGYKLSASAASLDPDIRRE
FAAVSRALDERFGRNAIIRDDKDLINRVPPVQRRAFEAMQERLRILQKAVRMKSSEQIISERRQRAVSRG
GGIDL
MAIMFVRAQVISRGVGRSIVSAAAYRHRARMMDEQAGTSFSYRGGRAELVHEELALPTQIPEWLRTAIDG
RTVAAASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSEGMVADWVYHDKDG
NPHIHLMTALRPLTQEGFGRKKVAVTGEDGAPLRVVTPDRPKGRIVYKLWAGDKETMKAWKIAWAETANR
HLALAGHEIRIDGRSYAEQGLDGIAQRHLGPEKAALSRRGATMYFAPADLARRQEMADRLLAEPEFLLKR
LGNERSTFDERDIAKALHRYVDDPSDFANIRAGLMASDDLVILKPQEVDPETGKVAEPAVFTTRDILRVE
YDMARSAQVLSERRGFAVSAESVAAAIENVETADPEKSFRLDPEQVDAVRHVAGDNGIAAVVGLAGAGKS
TLLAAARVAWESGGRRVIGAALAGKAAEGLEDSSGISSRTLAAWELIWANGHETLHRGDVLVIDEAGMVS
SQQMARVLKLVEQARAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREEWAREASRLFARGE
IEKGLDAYAGHGHLVEAETRAETIDRIVADWTEARKQAIRNSTVVANDGRLRGDALLVLAHTNQDVKRLN
EALRSVMTGEGALGEGRSFRTERGAREFAAGDRMIFLENARFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GDTLLTVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVERTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEAKPEWGRKPHVDHAAGVTGELVETGQAKFRPNDEDADDSPYADVKTDDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVEKVKVQVPIVDEQTGQKRFEEREVDRNVWTAKRIETAAERQERIERESHRP
DVFKQLVERLSRSGAKTTTLDFESEAGYRAYADDFARRRGLDHLSLAAAGVEESLSRRWAGIAERREQLA
KLWERASMALGFTIERERRVAYSEERAEPTPVLEADVVSGRYLIPPATGFTRSVEEDARLAQLASPAGKE
REAILRPLLEKIYRDPDAALVALNALSSDASIEPRRLVDDLAATPDRLGRLRGSDLIVDGRAARDELNAA
ITALTELLPLARAHATEFRRNGERFDLGEQQRRTHMSLPIPALSEKAMARLTEIEAVRERGGDDAYKTAF
ALVTEDRSIVQEIKAVSEALTARFGWSAFTPNADAVAERNMVERMPEDLASIRGEKLTRLFEAVKRFADE
QHLAERRNRSKIVAAASADQGMGTDKENAAVLPMLAAVTDFKTPVDEEARSRVLSAPLYRQQRAALAEAA
RVIWRDPAGAVGKIEELLAKGFAGERIAAAVTNDPAAYGALRGSDRLMDRMLAAGRERKDAVQAMPEAAA
RLRALGSAYVNVLDGERRAIAEERRRMTVAIPGLSKAAEEVLMRLTAEAKNNGYKLSASAASLDPDIRRE
FAAVSRALDERFGRNAIIRDDKDLINRVPPVQRRAFEAMQERLRILQKAVRMKSSEQIISERRQRAVSRG
GGIDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15723 | GenBank | WP_231285545 |
| Name | traG_RlegWSM1455_RS27740_pWSM1455_1 |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70501.42 Da Isoelectric Point: 9.2552
>WP_231285545.1 Ti-type conjugative transfer system protein TraG [Rhizobium laguerreae]
MALKCRLHPGLLLVLVPVAVTAIAVYITGWRWSALAHGLSGKSEYWFMRAAPVPALLFGPLTGLLTVWAL
PLHRRRPIAIASLFCFLAIAGFYALREYGRLAPSVESGTVSWDRALSYLDMVAVIGALAGFMVTAVAARI
SVVVPEQVKRAKGGTFGDADWLSMSAAAKLFPPDGEIVVGERYRVDKEVVYQLPFDPNDPSTWGRGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVLEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSESAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNTFRSSDIIAGGTDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGRFSRRALFMLDEVDLLGYIRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFVSYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKRGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHRPIRCGRAIYFRRKDMNAAAK
ANRFVKPVP
MALKCRLHPGLLLVLVPVAVTAIAVYITGWRWSALAHGLSGKSEYWFMRAAPVPALLFGPLTGLLTVWAL
PLHRRRPIAIASLFCFLAIAGFYALREYGRLAPSVESGTVSWDRALSYLDMVAVIGALAGFMVTAVAARI
SVVVPEQVKRAKGGTFGDADWLSMSAAAKLFPPDGEIVVGERYRVDKEVVYQLPFDPNDPSTWGRGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVLEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSESAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNTFRSSDIIAGGTDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGRFSRRALFMLDEVDLLGYIRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFVSYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKRGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHRPIRCGRAIYFRRKDMNAAAK
ANRFVKPVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 420845..430452
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RlegWSM1455_RS25930 (RlegWSM1455_25930) | 416654..417238 | - | 585 | WP_077980522 | TadE/TadG family type IV pilus assembly protein | - |
| RlegWSM1455_RS25935 (RlegWSM1455_25935) | 417240..417770 | - | 531 | WP_131640974 | TadE/TadG family type IV pilus assembly protein | - |
| RlegWSM1455_RS25940 (RlegWSM1455_25940) | 417767..419128 | - | 1362 | WP_131640973 | TadE/TadG family type IV pilus assembly protein | - |
| RlegWSM1455_RS25945 (RlegWSM1455_25945) | 419854..420207 | - | 354 | WP_077980519 | MarR family transcriptional regulator | - |
| RlegWSM1455_RS25950 (RlegWSM1455_25950) | 420299..420841 | + | 543 | WP_131640972 | hypothetical protein | - |
| RlegWSM1455_RS25955 (RlegWSM1455_25955) | 420845..421507 | + | 663 | WP_231285670 | lytic transglycosylase domain-containing protein | virB1 |
| RlegWSM1455_RS25960 (RlegWSM1455_25960) | 421504..421803 | + | 300 | WP_231285671 | TrbC/VirB2 family protein | virB2 |
| RlegWSM1455_RS25965 (RlegWSM1455_25965) | 421808..422146 | + | 339 | WP_231285673 | type IV secretion system protein VirB3 | virB3 |
| RlegWSM1455_RS25970 (RlegWSM1455_25970) | 422139..424505 | + | 2367 | WP_231285674 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| RlegWSM1455_RS25975 (RlegWSM1455_25975) | 424538..425203 | + | 666 | WP_231285725 | P-type DNA transfer protein VirB5 | virB5 |
| RlegWSM1455_RS25980 (RlegWSM1455_25980) | 425200..425433 | + | 234 | WP_231285675 | EexN family lipoprotein | - |
| RlegWSM1455_RS25985 (RlegWSM1455_25985) | 425437..426357 | + | 921 | WP_131663326 | type IV secretion system protein | virB6 |
| RlegWSM1455_RS25990 (RlegWSM1455_25990) | 426410..426694 | + | 285 | WP_231285676 | hypothetical protein | - |
| RlegWSM1455_RS25995 (RlegWSM1455_25995) | 426696..427367 | + | 672 | WP_221959625 | virB8 family protein | virB8 |
| RlegWSM1455_RS26000 (RlegWSM1455_26000) | 427364..428221 | + | 858 | WP_231285677 | P-type conjugative transfer protein VirB9 | virB9 |
| RlegWSM1455_RS26005 (RlegWSM1455_26005) | 428231..429403 | + | 1173 | WP_231285679 | type IV secretion system protein VirB10 | virB10 |
| RlegWSM1455_RS26010 (RlegWSM1455_26010) | 429412..430452 | + | 1041 | WP_231285682 | P-type DNA transfer ATPase VirB11 | virB11 |
| RlegWSM1455_RS26015 (RlegWSM1455_26015) | 430608..430823 | - | 216 | WP_231285683 | DUF3072 domain-containing protein | - |
| RlegWSM1455_RS26020 (RlegWSM1455_26020) | 430954..431862 | - | 909 | WP_231285684 | nucleotidyltransferase and HEPN domain-containing protein | - |
| RlegWSM1455_RS26025 (RlegWSM1455_26025) | 432277..432519 | + | 243 | WP_222023730 | type II toxin-antitoxin system Phd/YefM family antitoxin | - |
| RlegWSM1455_RS26030 (RlegWSM1455_26030) | 432519..432953 | + | 435 | WP_231285685 | type II toxin-antitoxin system VapC family toxin | - |
| RlegWSM1455_RS26035 (RlegWSM1455_26035) | 433042..434188 | - | 1147 | Protein_423 | site-specific integrase | - |
Host bacterium
| ID | 21675 | GenBank | NZ_CP088091 |
| Plasmid name | pWSM1455_1 | Incompatibility group | - |
| Plasmid size | 1064136 bp | Coordinate of oriT [Strand] | 744893..744921 [+] |
| Host baterium | Rhizobium laguerreae strain WSM1455 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nifN, nifE, nifK, nifD, nifH, nodM, nodE, nodF, nodD, nodA, nodC, nodI, nodJ, nifB, fixX, fixC, fixB, fixA, fixN, fixO, fixQ, fixG |
| Anti-CRISPR | AcrIIA7 |