Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121222 |
| Name | oriT_pLU4 |
| Organism | Limosilactobacillus reuteri strain LU4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KM063576 (3222..3259 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 1..7, 19..25 (ACACAAC..GTTGTGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pLU4
ACACAACCAACTTTAGTTGTTGTGTGTAAGTGCGCATT
ACACAACCAACTTTAGTTGTTGTGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file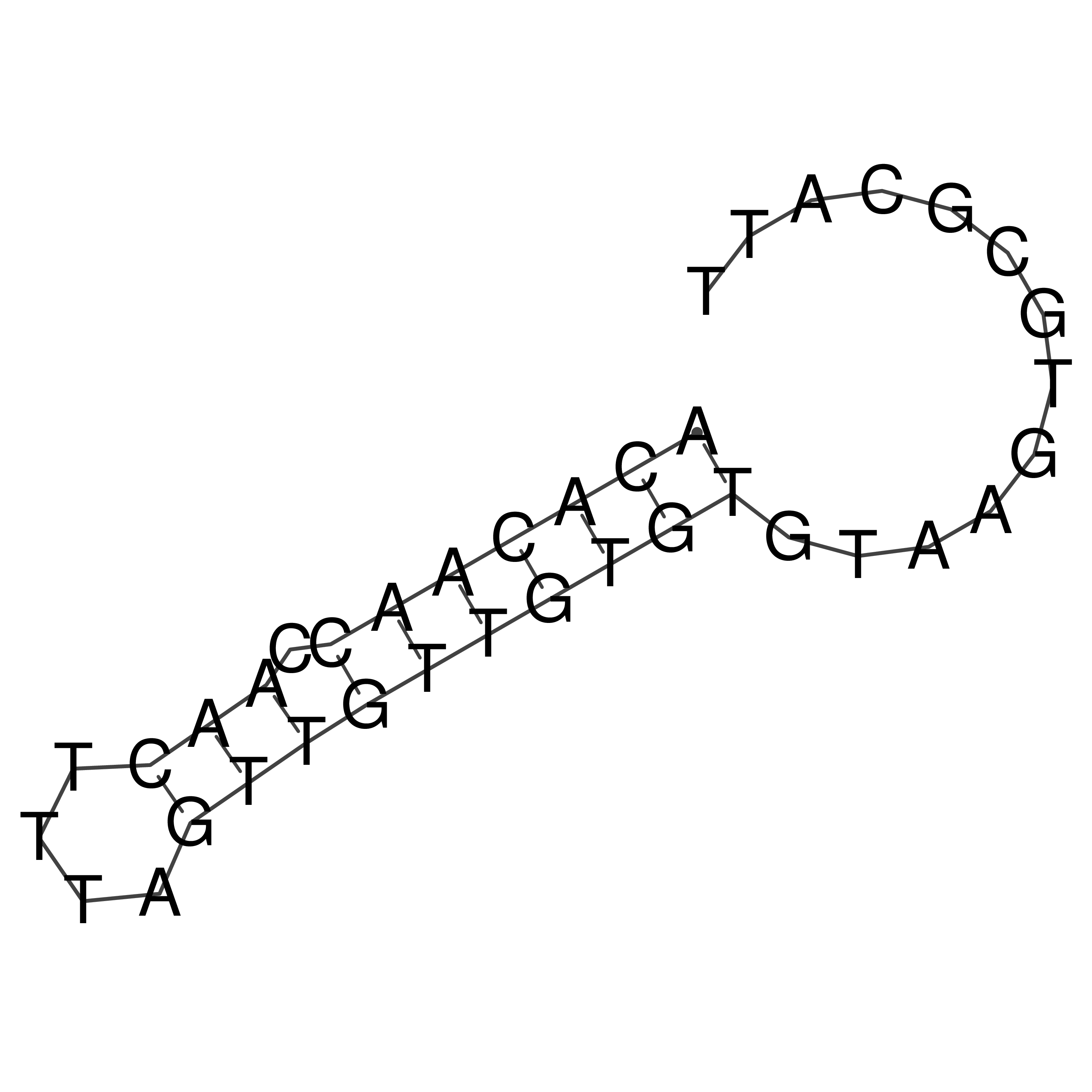
Relaxase
| ID | 13404 | GenBank | WP_075667750 |
| Name | mobQ_HTD43_RS00035_pLU4 |
UniProt ID | A0A1L7GYD1 |
| Length | 687 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 687 a.a. Molecular weight: 80874.51 Da Isoelectric Point: 9.5890
>WP_075667750.1 MULTISPECIES: MobQ family relaxase [Limosilactobacillus]
MAIFHMSFSNISAGKGRSAIASAAYRSGKKLFDQKEGRSYFYARSVMPESFILTPKNAPEWASDREKLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQKNFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNQDGTWGLKSKRENILDENGNKTYTGNSRFPRSRKVWLVDWDKKEKINQWRHNWAVSVNQVLEQKNIP
DRISEKSFIEQGIDDTPMQHEGINSKRYERKEFNQQVKNYRKAKASYKNNQEKVINRGHLDSLSKHFSFN
EKRVVKELSHELKTYISLESLDDKRRMLFNWKNSTLIKHAVGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETDSEQTIFKGSELNERLMNIRDDLLTQQVLTFTKRPYVGWKLL
MQQEKEVKIELKYTLMIHDDSLESLEHVDQGLLEKYSPTEQQKITRAVKDLRTIMAVKQVIQTQYQEVLR
RAFPNGDFNELPMIKQEQAYTAVMYYDPALKPCKAETIAQWQENPPRVFNTQEHLQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKVDPTIKNSQIEKIQKQLKEQQAKDDQYRKVNMGHYQPLNYKPVSPSY
YLKTAFSNAIMTALYAHDEDYERQKQARSLKETEWEMTKKQRQHQTRNRHEDGGMHL
MAIFHMSFSNISAGKGRSAIASAAYRSGKKLFDQKEGRSYFYARSVMPESFILTPKNAPEWASDREKLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQKNFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNQDGTWGLKSKRENILDENGNKTYTGNSRFPRSRKVWLVDWDKKEKINQWRHNWAVSVNQVLEQKNIP
DRISEKSFIEQGIDDTPMQHEGINSKRYERKEFNQQVKNYRKAKASYKNNQEKVINRGHLDSLSKHFSFN
EKRVVKELSHELKTYISLESLDDKRRMLFNWKNSTLIKHAVGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETDSEQTIFKGSELNERLMNIRDDLLTQQVLTFTKRPYVGWKLL
MQQEKEVKIELKYTLMIHDDSLESLEHVDQGLLEKYSPTEQQKITRAVKDLRTIMAVKQVIQTQYQEVLR
RAFPNGDFNELPMIKQEQAYTAVMYYDPALKPCKAETIAQWQENPPRVFNTQEHLQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKVDPTIKNSQIEKIQKQLKEQQAKDDQYRKVNMGHYQPLNYKPVSPSY
YLKTAFSNAIMTALYAHDEDYERQKQARSLKETEWEMTKKQRQHQTRNRHEDGGMHL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1L7GYD1 |
T4CP
| ID | 15708 | GenBank | WP_075667768 |
| Name | t4cp2_HTD43_RS00095_pLU4 |
UniProt ID | _ |
| Length | 516 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 516 a.a. Molecular weight: 58092.27 Da Isoelectric Point: 4.8806
>WP_075667768.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Limosilactobacillus]
MNGTILGIVDKQIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRSPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMTDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVADFTSFSDFDLKEIGNDKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKEKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEDQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPEASQETNFKNKVEKWKAKLKETAQKEVANKMSQKEEEQQQDNL
DSDLERVSNKDSQTEAPEEDDNNLFG
MNGTILGIVDKQIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRSPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMTDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVADFTSFSDFDLKEIGNDKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKEKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEDQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPEASQETNFKNKVEKWKAKLKETAQKEVANKMSQKEEEQQQDNL
DSDLERVSNKDSQTEAPEEDDNNLFG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6810..16898
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTD43_RS00020 (LRELU4_p004) | 1892..2572 | + | 681 | WP_172686338 | zeta toxin family protein | - |
| HTD43_RS00025 (LRELU4_p005) | 2562..2840 | - | 279 | WP_172686339 | hypothetical protein | - |
| HTD43_RS00030 (LRELU4_p006) | 2863..3090 | - | 228 | WP_075667776 | hypothetical protein | - |
| HTD43_RS00035 (LRELU4_p007) | 3343..5406 | + | 2064 | WP_075667750 | MobQ family relaxase | - |
| HTD43_RS00040 (LRELU4_p008) | 5490..5801 | + | 312 | WP_003712158 | hypothetical protein | - |
| HTD43_RS00045 (LRELU4_p009) | 5838..6452 | + | 615 | WP_075667749 | hypothetical protein | - |
| HTD43_RS00050 (LRELU4_p010) | 6454..6789 | + | 336 | WP_003590767 | CagC family type IV secretion system protein | - |
| HTD43_RS00055 (LRELU4_p011) | 6810..7172 | + | 363 | WP_003590770 | conjugal transfer protein | trsC |
| HTD43_RS00060 (LRELU4_p012) | 7141..7800 | + | 660 | WP_003712179 | TrsD/TraD family conjugative transfer protein | trsD |
| HTD43_RS00065 (LRELU4_p013) | 7812..9830 | + | 2019 | WP_075667775 | VirB4 family type IV secretion system protein | virb4 |
| HTD43_RS00070 (LRELU4_p014) | 9823..11241 | + | 1419 | WP_172686340 | conjugal transfer protein | trsF |
| HTD43_RS00075 (LRELU4_p015) | 11242..12399 | + | 1158 | WP_172686341 | phage tail tip lysozyme | prgK |
| HTD43_RS00080 (LRELU4_p016) | 12413..13030 | + | 618 | WP_075667772 | hypothetical protein | - |
| HTD43_RS00085 (LRELU4_p017) | 13017..13385 | + | 369 | WP_075667771 | thioredoxin family protein | - |
| HTD43_RS00090 (LRELU4_p018) | 13386..13856 | + | 471 | WP_075667770 | conjugal transfer protein | trsJ |
| HTD43_RS00095 (LRELU4_p020) | 14086..15636 | + | 1551 | WP_075667768 | VirD4-like conjugal transfer protein, CD1115 family | - |
| HTD43_RS00100 (LRELU4_p021) | 15651..16040 | + | 390 | WP_075667767 | hypothetical protein | - |
| HTD43_RS00105 (LRELU4_p022) | 16059..16898 | + | 840 | WP_075667766 | conjugal transfer protein TrbL family protein | trsL |
| HTD43_RS00110 (LRELU4_p023) | 16913..17323 | + | 411 | WP_075667765 | hypothetical protein | - |
| HTD43_RS00115 (LRELU4_p024) | 17330..19465 | + | 2136 | WP_172686342 | type IA DNA topoisomerase | - |
| HTD43_RS00120 (LRELU4_p025) | 19585..19800 | + | 216 | WP_075667763 | hypothetical protein | - |
| HTD43_RS00125 (LRELU4_p026) | 19803..20927 | + | 1125 | WP_172686343 | ArdC-like ssDNA-binding domain-containing protein | - |
| HTD43_RS00130 | 21081..21170 | - | 90 | WP_021353390 | putative holin-like toxin | - |
Host bacterium
| ID | 21650 | GenBank | NZ_KM063576 |
| Plasmid name | pLU4 | Incompatibility group | - |
| Plasmid size | 33411 bp | Coordinate of oriT [Strand] | 3222..3259 [-] |
| Host baterium | Limosilactobacillus reuteri strain LU4 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |