Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121215 |
| Name | oriT_pLM159.2 |
| Organism | Xanthomonas vesicatoria strain LM159 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP018471 (45117..45215 [-], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
>oriT_pLM159.2
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file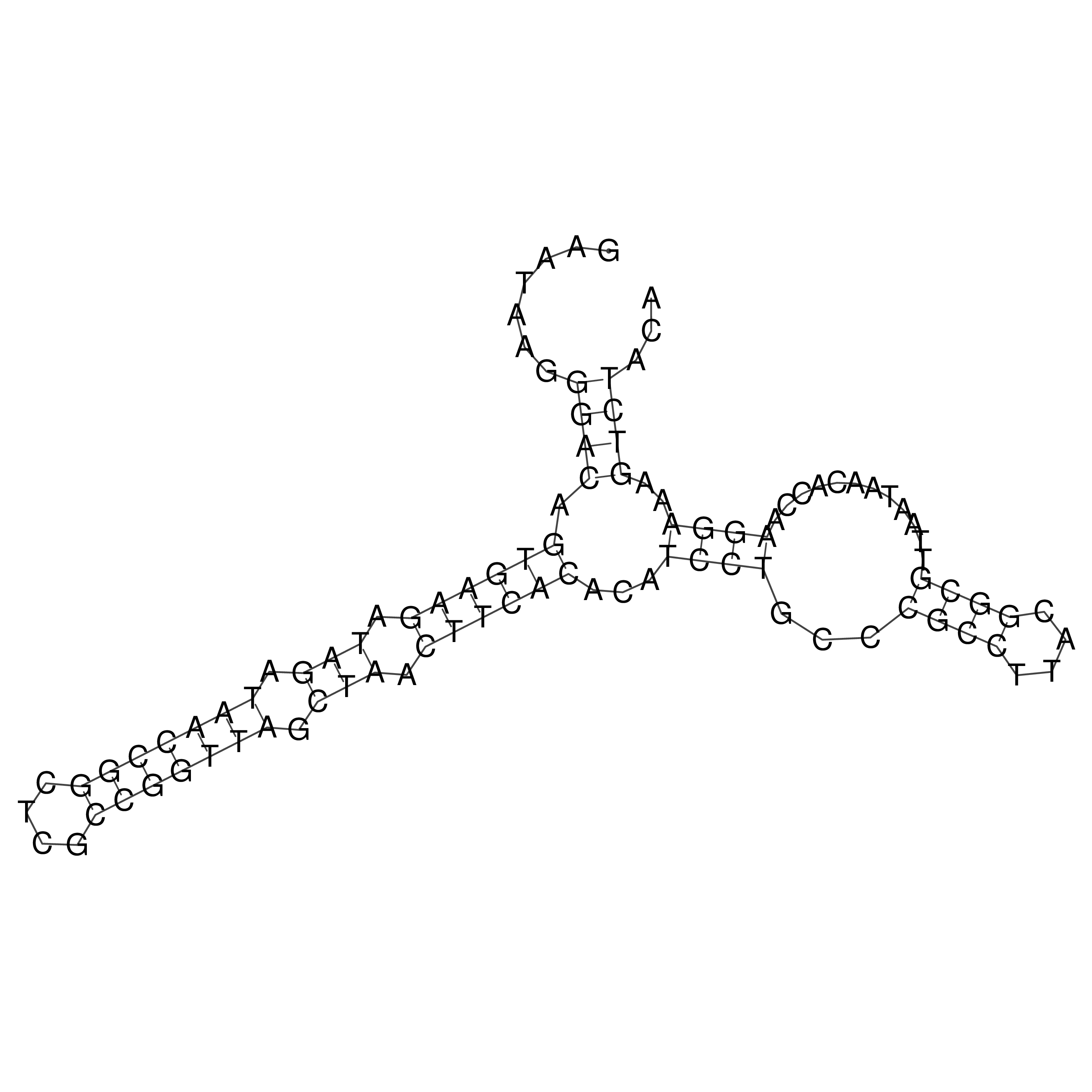
Relaxase
| ID | 13398 | GenBank | WP_058664035 |
| Name | traI_BI313_RS23735_pLM159.2 |
UniProt ID | _ |
| Length | 746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 746 a.a. Molecular weight: 81930.51 Da Isoelectric Point: 10.7900
>WP_058664035.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Pseudomonadota]
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGQVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTTLERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQSLTTARGQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRTNKKLLYAQASKALRSE
IQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGSADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRAGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAASVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVSRHVEQQGTQ
PDHALRRGISRPGAGVGPTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALSFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGQVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTTLERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQSLTTARGQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRTNKKLLYAQASKALRSE
IQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGSADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRAGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAASVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVSRHVEQQGTQ
PDHALRRGISRPGAGVGPTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALSFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15701 | GenBank | WP_074052969 |
| Name | t4cp2_BI313_RS23445_pLM159.2 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 93902.02 Da Isoelectric Point: 6.3901
>WP_074052969.1 MULTISPECIES: VirB4 family type IV secretion/conjugal transfer ATPase [Gammaproteobacteria]
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSSAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKLT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSSAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKLT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15702 | GenBank | WP_074052986 |
| Name | t4cp2_BI313_RS23740_pLM159.2 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69925.14 Da Isoelectric Point: 8.2993
>WP_074052986.1 type IV secretory system conjugative DNA transfer family protein [Xanthomonas vesicatoria]
MKIKMNNAVGPQVRSAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDEGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRVVAQAET
EGEGITI
MKIKMNNAVGPQVRSAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDEGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRVVAQAET
EGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4024..13731
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BI313_RS23420 (BI313_23320) | 30..2915 | - | 2886 | WP_001138082 | Tn3 family transposase | - |
| BI313_RS23425 (BI313_23325) | 3041..3142 | + | 102 | Protein_1 | recombinase family protein | - |
| BI313_RS23430 (BI313_23330) | 3197..3961 | - | 765 | WP_001389365 | IS6-like element IS6100 family transposase | - |
| BI313_RS23435 (BI313_23335) | 4024..4488 | + | 465 | WP_074052968 | conjugal transfer system pilin TrbC | virB2 |
| BI313_RS23440 (BI313_23340) | 4492..4803 | + | 312 | WP_001207961 | conjugal transfer protein TrbD | virB3 |
| BI313_RS23445 (BI313_23345) | 4800..7358 | + | 2559 | WP_074052969 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| BI313_RS23450 (BI313_23350) | 7355..8137 | + | 783 | WP_011798705 | conjugal transfer protein TrbF | virB8 |
| BI313_RS23455 (BI313_23355) | 8155..9054 | + | 900 | WP_000753228 | P-type conjugative transfer protein TrbG | virB9 |
| BI313_RS23460 (BI313_23360) | 9057..9545 | + | 489 | WP_074052970 | conjugal transfer protein TrbH | - |
| BI313_RS23465 (BI313_23365) | 9550..10968 | + | 1419 | WP_011798703 | TrbI/VirB10 family protein | virB10 |
| BI313_RS23470 (BI313_23370) | 10986..11750 | + | 765 | WP_011798702 | P-type conjugative transfer protein TrbJ | virB5 |
| BI313_RS23475 (BI313_23375) | 11760..11987 | + | 228 | WP_011798701 | entry exclusion lipoprotein TrbK | - |
| BI313_RS23480 (BI313_23380) | 11998..13731 | + | 1734 | WP_011798700 | P-type conjugative transfer protein TrbL | virB6 |
| BI313_RS23485 (BI313_23385) | 13748..14332 | + | 585 | WP_011798699 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| BI313_RS23490 (BI313_23390) | 14346..14990 | + | 645 | WP_031942725 | transglycosylase SLT domain-containing protein | - |
| BI313_RS23495 (BI313_23395) | 14987..15253 | + | 267 | WP_074052971 | hypothetical protein | - |
| BI313_RS23500 (BI313_23400) | 15330..15965 | + | 636 | WP_229006823 | TraX family protein | - |
| BI313_RS23505 (BI313_23405) | 15981..16412 | + | 432 | WP_074052990 | OmpA family protein | - |
| BI313_RS23510 (BI313_23410) | 16569..17243 | + | 675 | WP_074052972 | SAM-dependent methyltransferase | - |
| BI313_RS23515 (BI313_23415) | 17245..18096 | - | 852 | WP_231117434 | recombinase family protein | - |
Host bacterium
| ID | 21643 | GenBank | NZ_CP018471 |
| Plasmid name | pLM159.2 | Incompatibility group | IncP1 |
| Plasmid size | 62784 bp | Coordinate of oriT [Strand] | 45117..45215 [-] |
| Host baterium | Xanthomonas vesicatoria strain LM159 |
Cargo genes
| Drug resistance gene | aph(6)-Id, aph(3'')-Ib |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |