Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121214 |
| Name | oriT_pT136b |
| Organism | Rhizobium sp. T136 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP088011 (467321..467353 [+], 33 nt) |
| oriT length | 33 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 33 nt
>oriT_pT136b
TCCAAGGGCGCAATTATACGTCGCTGGCGCGAC
TCCAAGGGCGCAATTATACGTCGCTGGCGCGAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file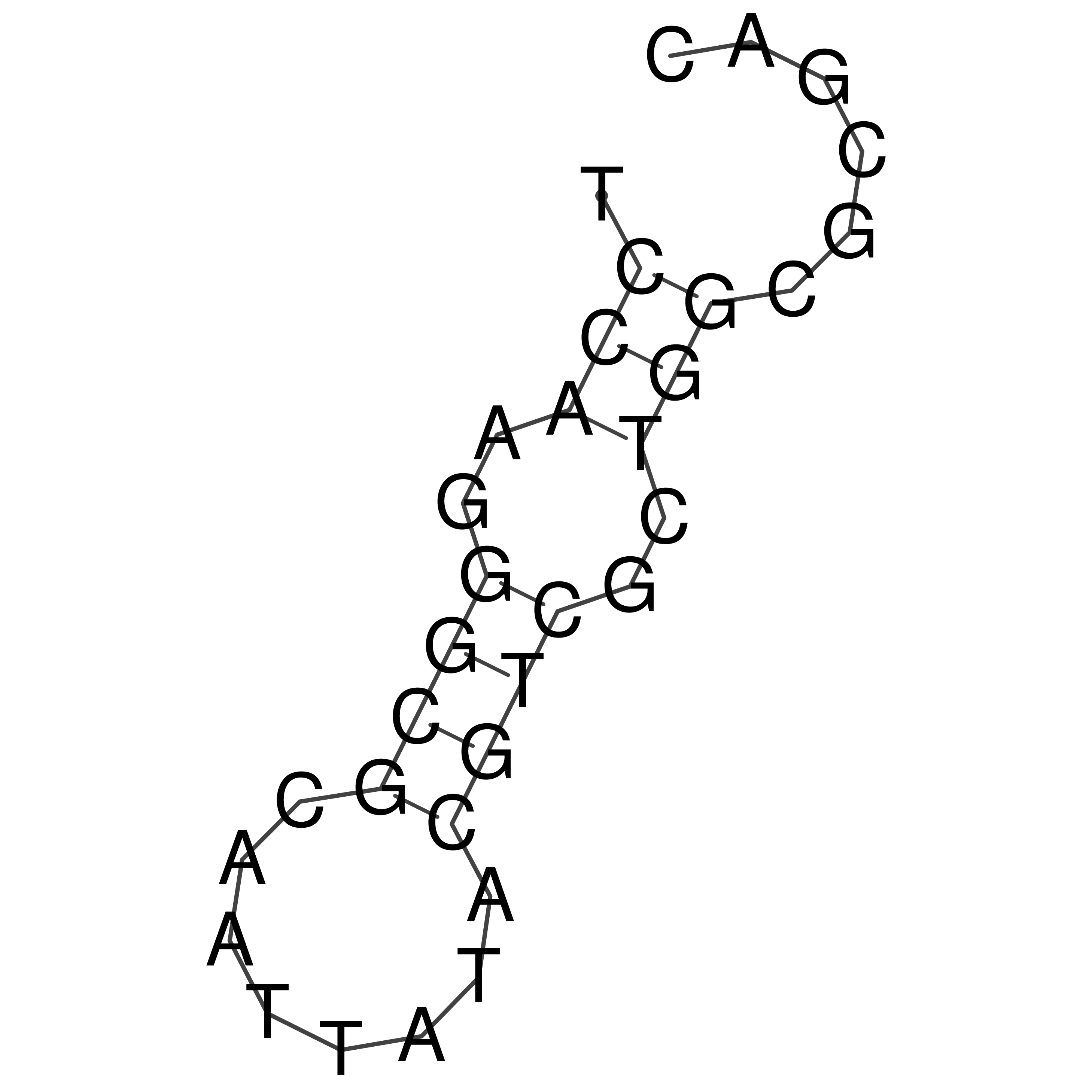
Relaxase
| ID | 13397 | GenBank | WP_024317491 |
| Name | traA_LPB79_RS32570_pT136b |
UniProt ID | W6S1J9 |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122775.11 Da Isoelectric Point: 10.1192
>WP_024317491.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEELVVPSDAPEWLRMMVADR
SVAGASEAFWNKVEVFEKRSDAQLAKDVTIALPIELTAAQNIALVRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLAPDGKPIRNDAGKIVYELWAGSAEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFERQGIALEPTIHLGVGTKAIERKAEQTDRKQETPASKLERVKFQEARRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDASLFQSLMVRILQSPEALRLERERINFATGIRTPAKYTSRQM
IRLEAEMVNRAIWLSGRASHGVREAVLEATFARHARLSVEQRTAIEHVAGGERIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNEGRNHIDDKTVFVLDEAGMVSSRQM
ALFVDAVTKAGAKLVLVGDPEQLQPIGAGAGFRAIADRIGYAELETIYRQRQQWMRDASLDLARGKVGEA
VEAYRAHGRFNGLDLKAQAVESLIADWNHGYDPSKTTLILAHLRRDVRMLNEMARAKLVERGIIADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRLVAELGGGEHRRLVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KASFYRQALRFAEARGLHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAMGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSFGQAVEDKLAADPGLKKQWEDVSTRFHLVYAQPEAAFNAINVDAMAKDQSVAQS
TLERIAGAPESFGALKGKTGLLASRADKQEREKAVANVPALARNLKRYLRERAEAEFKHETEERAVRLKV
SVDIPALSASAKQTLERVRDAIDSNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTSG
KTFETVTSGMTVSQKVEVQSAWNSMRTVQQLGSHERTTEALKQAETLRQTKSQGRSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEELVVPSDAPEWLRMMVADR
SVAGASEAFWNKVEVFEKRSDAQLAKDVTIALPIELTAAQNIALVRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLAPDGKPIRNDAGKIVYELWAGSAEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFERQGIALEPTIHLGVGTKAIERKAEQTDRKQETPASKLERVKFQEARRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDASLFQSLMVRILQSPEALRLERERINFATGIRTPAKYTSRQM
IRLEAEMVNRAIWLSGRASHGVREAVLEATFARHARLSVEQRTAIEHVAGGERIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNEGRNHIDDKTVFVLDEAGMVSSRQM
ALFVDAVTKAGAKLVLVGDPEQLQPIGAGAGFRAIADRIGYAELETIYRQRQQWMRDASLDLARGKVGEA
VEAYRAHGRFNGLDLKAQAVESLIADWNHGYDPSKTTLILAHLRRDVRMLNEMARAKLVERGIIADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRLVAELGGGEHRRLVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KASFYRQALRFAEARGLHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAMGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSFGQAVEDKLAADPGLKKQWEDVSTRFHLVYAQPEAAFNAINVDAMAKDQSVAQS
TLERIAGAPESFGALKGKTGLLASRADKQEREKAVANVPALARNLKRYLRERAEAEFKHETEERAVRLKV
SVDIPALSASAKQTLERVRDAIDSNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTSG
KTFETVTSGMTVSQKVEVQSAWNSMRTVQQLGSHERTTEALKQAETLRQTKSQGRSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W6S1J9 |
T4CP
| ID | 15699 | GenBank | WP_024317494 |
| Name | traG_LPB79_RS32555_pT136b |
UniProt ID | _ |
| Length | 659 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 659 a.a. Molecular weight: 70953.33 Da Isoelectric Point: 9.2056
>WP_024317494.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVVVPTALMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGVLLLFASAGSI
NIKQAGWGVVAGSSGTILVAAVRETMRLSAVIKAAPADKTVLAFLDPATSIGASAALLCACFALRVALIG
NAAFAGAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVASQAFRADSAEVWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPIVIAHRGGAGRDVFVLN
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHMDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFPTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRDTYGGRDAASKWFESASWISFAAINDPETADYISKRCGMTT
VEIDQVSRSFHAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRVCIG
ENRFHSMGGREKEASNATPAVNPYEPNRL
MTINRIALVVVPTALMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGVLLLFASAGSI
NIKQAGWGVVAGSSGTILVAAVRETMRLSAVIKAAPADKTVLAFLDPATSIGASAALLCACFALRVALIG
NAAFAGAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVASQAFRADSAEVWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPIVIAHRGGAGRDVFVLN
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHMDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFPTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRDTYGGRDAASKWFESASWISFAAINDPETADYISKRCGMTT
VEIDQVSRSFHAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRVCIG
ENRFHSMGGREKEASNATPAVNPYEPNRL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15700 | GenBank | WP_024317476 |
| Name | t4cp2_LPB79_RS32645_pT136b |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91872.73 Da Isoelectric Point: 5.8129
>WP_024317476.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKRFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDFERNELSRRINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREKGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFDNTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMRMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAVRRLIQAEGYGARIETLNATDAYLGSLPGNWYCNIREPLINTCNLADLIPLNSVWP
GNPVAPCPFFAPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFLRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNVEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDYRN
AISRQIGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEHLMNLG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGRDWPIHWLETRGVHDAASLLKCE
MVALKRFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDFERNELSRRINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREKGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFDNTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMRMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAVRRLIQAEGYGARIETLNATDAYLGSLPGNWYCNIREPLINTCNLADLIPLNSVWP
GNPVAPCPFFAPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFLRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNVEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDYRN
AISRQIGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEHLMNLG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGRDWPIHWLETRGVHDAASLLKCE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 472492..485410
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LPB79_RS32575 (LPB79_32575) | 470825..471322 | + | 498 | WP_231052413 | conjugative transfer signal peptidase TraF | - |
| LPB79_RS32580 (LPB79_32580) | 471312..472475 | + | 1164 | WP_024317489 | conjugal transfer protein TraB | - |
| LPB79_RS32585 (LPB79_32585) | 472492..473115 | + | 624 | WP_024317488 | TraH family protein | virB1 |
| LPB79_RS32590 (LPB79_32590) | 473371..473913 | - | 543 | WP_024317487 | hypothetical protein | - |
| LPB79_RS32595 (LPB79_32595) | 474037..474276 | - | 240 | WP_024317486 | helix-turn-helix domain-containing protein | - |
| LPB79_RS32600 (LPB79_32600) | 474567..474857 | + | 291 | WP_024317485 | transcriptional repressor TraM | - |
| LPB79_RS32605 (LPB79_32605) | 474861..475565 | - | 705 | WP_024317484 | autoinducer binding domain-containing protein | - |
| LPB79_RS32610 (LPB79_32610) | 475890..477188 | - | 1299 | WP_024317483 | IncP-type conjugal transfer protein TrbI | virB10 |
| LPB79_RS32615 (LPB79_32615) | 477200..477640 | - | 441 | WP_024317482 | conjugal transfer protein TrbH | - |
| LPB79_RS32620 (LPB79_32620) | 477644..478456 | - | 813 | WP_024317481 | P-type conjugative transfer protein TrbG | virB9 |
| LPB79_RS32625 (LPB79_32625) | 478474..479136 | - | 663 | WP_024317480 | conjugal transfer protein TrbF | virB8 |
| LPB79_RS32630 (LPB79_32630) | 479157..480338 | - | 1182 | WP_024317479 | P-type conjugative transfer protein TrbL | virB6 |
| LPB79_RS32635 (LPB79_32635) | 480332..480529 | - | 198 | WP_024317478 | entry exclusion protein TrbK | - |
| LPB79_RS32640 (LPB79_32640) | 480526..481329 | - | 804 | WP_024317477 | P-type conjugative transfer protein TrbJ | virB5 |
| LPB79_RS32645 (LPB79_32645) | 481301..483757 | - | 2457 | WP_024317476 | conjugal transfer protein TrbE | virb4 |
| LPB79_RS32650 (LPB79_32650) | 483768..484067 | - | 300 | WP_024317475 | conjugal transfer protein TrbD | virB3 |
| LPB79_RS32655 (LPB79_32655) | 484060..484443 | - | 384 | WP_024317474 | TrbC/VirB2 family protein | virB2 |
| LPB79_RS32660 (LPB79_32660) | 484433..485410 | - | 978 | WP_024317473 | P-type conjugative transfer ATPase TrbB | virB11 |
| LPB79_RS32665 (LPB79_32665) | 485421..486044 | - | 624 | WP_024317472 | acyl-homoserine-lactone synthase TraI | - |
| LPB79_RS32670 (LPB79_32670) | 486733..487956 | + | 1224 | WP_024317471 | plasmid partitioning protein RepA | - |
| LPB79_RS32675 (LPB79_32675) | 487953..488990 | + | 1038 | WP_024317470 | plasmid partitioning protein RepB | - |
| LPB79_RS32680 (LPB79_32680) | 489182..490396 | + | 1215 | WP_024317469 | plasmid replication protein RepC | - |
Host bacterium
| ID | 21642 | GenBank | NZ_CP088011 |
| Plasmid name | pT136b | Incompatibility group | - |
| Plasmid size | 806677 bp | Coordinate of oriT [Strand] | 467321..467353 [+] |
| Host baterium | Rhizobium sp. T136 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | LinG, dxnH |
| Symbiosis gene | nodD, nodH, nodF, nodE, nodJ, nodI, nodC, nodB, nodA, fixU, nifZ, nifB, fixX, fixC, fixB, fixA, nifW, nifS, nifH, nifD, nifK, nifE, nifN, nifX, mLTONO_5203, fixN, fixO, fixQ, nodM |
| Anti-CRISPR | - |