Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121204 |
| Name | oriT_p34399-106.698kb |
| Organism | Enterobacter hormaechei subsp. xiangfangensis strain 34399 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP010385 (80344..80430 [-], 87 nt) |
| oriT length | 87 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 87 nt
>oriT_p34399-106.698kb
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTGATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTCTTTCG
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTGATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTCTTTCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file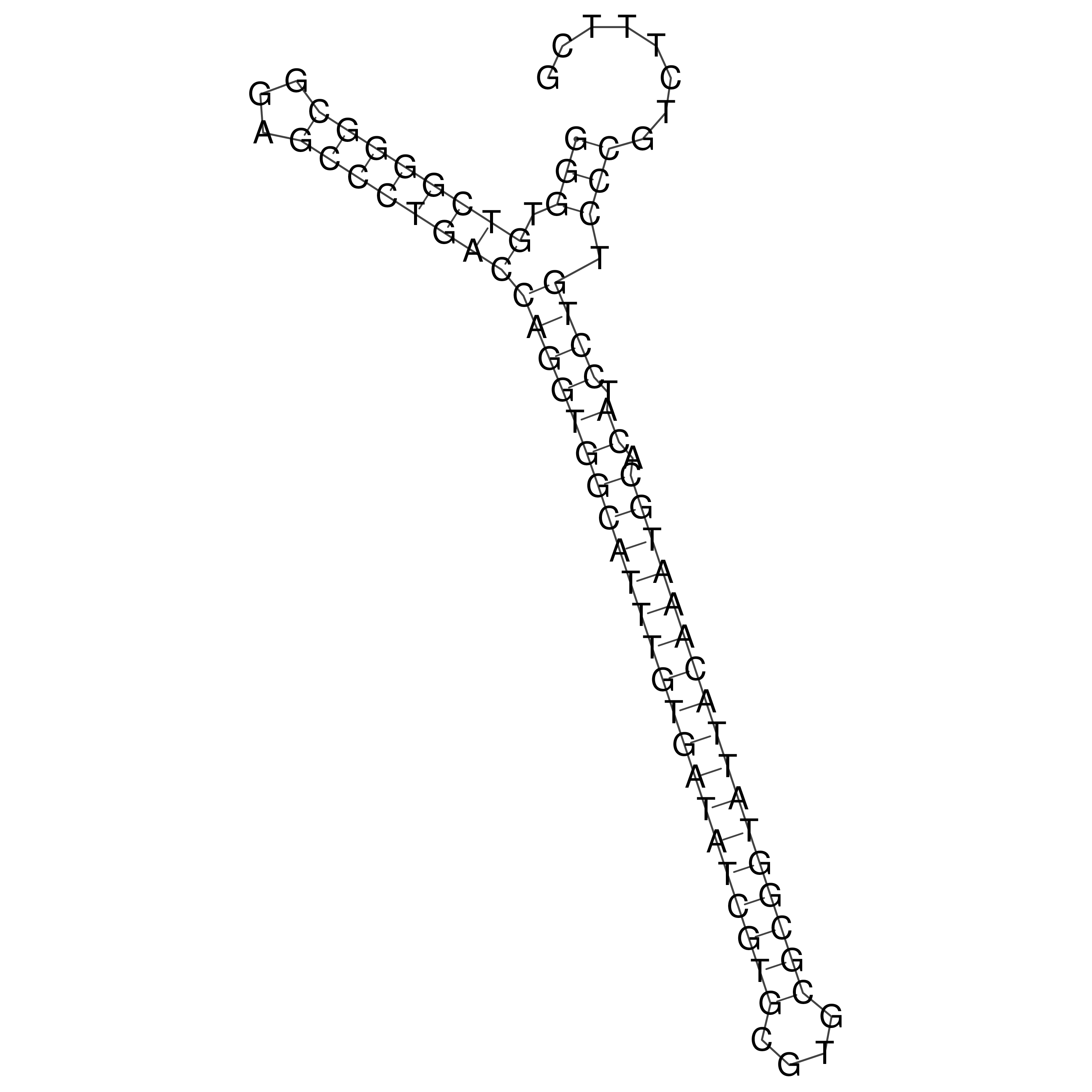
Relaxase
| ID | 13388 | GenBank | WP_022650008 |
| Name | Relaxase_LI66_RS23340_p34399-106.698kb |
UniProt ID | A0A4Q2QTK9 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 102314.37 Da Isoelectric Point: 8.2756
>WP_022650008.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacterales]
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVRASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFSARKTKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLAGHQFVAAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSVERDRQSAWKRGRNQSLREYIADTAIAGLRESPVTDWSSLHQRFAKEGLFLTKE
SGELKVKDGWDSERPGVTLSAFGHAFDTDKLIRKLGEFVPPVQDIFSQVPAVGRYEPDKIIMPSRPERMT
EKESLSDYAIARLRAPLIALDQDPTQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDSTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKDIFQQVPPAEQFTGGSLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDIESLIGSGEFTWERCHELLARQGLLLMREQQGLVVMDAYNHEQTPVKASHVHPDLTLAR
AEPHAGSFVSVPADIFERVKPASRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQAIHDECRIRKARIRIEHRDPLVRKLHYHIAELQRMQALIALKETMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKSTTDNRCVVICEPGGTPMYSGVPGLRASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNSGDYDALKRDMVKVAPVLFSRSPSVAMAPEGNNEQFNTAFAKM
VAWHNVNHRDEGEYQISRPEVDQVRVESEMRFAEMTNQGSNTEAVRNESGPNWKPPSPL
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVRASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFSARKTKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLAGHQFVAAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSVERDRQSAWKRGRNQSLREYIADTAIAGLRESPVTDWSSLHQRFAKEGLFLTKE
SGELKVKDGWDSERPGVTLSAFGHAFDTDKLIRKLGEFVPPVQDIFSQVPAVGRYEPDKIIMPSRPERMT
EKESLSDYAIARLRAPLIALDQDPTQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDSTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKDIFQQVPPAEQFTGGSLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDIESLIGSGEFTWERCHELLARQGLLLMREQQGLVVMDAYNHEQTPVKASHVHPDLTLAR
AEPHAGSFVSVPADIFERVKPASRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQAIHDECRIRKARIRIEHRDPLVRKLHYHIAELQRMQALIALKETMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKSTTDNRCVVICEPGGTPMYSGVPGLRASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNSGDYDALKRDMVKVAPVLFSRSPSVAMAPEGNNEQFNTAFAKM
VAWHNVNHRDEGEYQISRPEVDQVRVESEMRFAEMTNQGSNTEAVRNESGPNWKPPSPL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A4Q2QTK9 |
T4CP
| ID | 15688 | GenBank | WP_022650006 |
| Name | trbC_LI66_RS23350_p34399-106.698kb |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87660.94 Da Isoelectric Point: 6.8660
>WP_022650006.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacterales]
MNYTPVNQALMKRNAWNSPVLELLRDHLLYAGIMAGSVVAGFIWPLAIPLLLLLAIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSLFRRFPSLFQYETTQESKGKGIFYLGYRRMNDIGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IHNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAELYCRAIDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHRLE
GLDSEALEITKYKGPFPYMNYLDEVGAYYTERIAVQATQVRSLDFSIVMMSQDQERIESQTSATNVATLM
QNAGTKVAGKIVSDDKTAKTITNAAGQEARANMVSLQRRDGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAPENVSKIIG
VLTEKPSRRRKAAKAEPWKIIDTFQQRLANRQEAHNLMTEYDTDHRGREENLWEEALEVIRTTTMAERTI
RYITLNKPESEPTQVSAEEIQLAPDAILRTLTLPQPAYIPPSRYRNEPFISPKLPDSPHPDMEWPE
MNYTPVNQALMKRNAWNSPVLELLRDHLLYAGIMAGSVVAGFIWPLAIPLLLLLAIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSLFRRFPSLFQYETTQESKGKGIFYLGYRRMNDIGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IHNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAELYCRAIDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHRLE
GLDSEALEITKYKGPFPYMNYLDEVGAYYTERIAVQATQVRSLDFSIVMMSQDQERIESQTSATNVATLM
QNAGTKVAGKIVSDDKTAKTITNAAGQEARANMVSLQRRDGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAPENVSKIIG
VLTEKPSRRRKAAKAEPWKIIDTFQQRLANRQEAHNLMTEYDTDHRGREENLWEEALEVIRTTTMAERTI
RYITLNKPESEPTQVSAEEIQLAPDAILRTLTLPQPAYIPPSRYRNEPFISPKLPDSPHPDMEWPE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 608..14299
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LI66_RS22875 (LI66_22875) | 608..895 | - | 288 | WP_022649988 | hypothetical protein | traK |
| LI66_RS22880 (LI66_22880) | 892..2052 | - | 1161 | WP_022649987 | plasmid transfer ATPase TraJ | virB11 |
| LI66_RS22885 (LI66_22885) | 2065..2871 | - | 807 | WP_022649986 | type IV secretory system conjugative DNA transfer family protein | traI |
| LI66_RS22890 (LI66_22890) | 2868..3341 | - | 474 | WP_022649985 | DotD/TraH family lipoprotein | - |
| LI66_RS25905 | 3412..3828 | - | 417 | WP_123839394 | hypothetical protein | - |
| LI66_RS22895 (LI66_22895) | 3937..5142 | - | 1206 | WP_022649984 | conjugal transfer protein TraF | - |
| LI66_RS24485 | 5248..6084 | - | 837 | WP_022649983 | hypothetical protein | traE |
| LI66_RS25910 | 6231..6707 | - | 477 | WP_022649982 | hypothetical protein | - |
| LI66_RS22905 (LI66_22905) | 6727..7887 | - | 1161 | WP_022649981 | site-specific integrase | - |
| LI66_RS22910 (LI66_22910) | 8465..9802 | - | 1338 | WP_040110252 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| LI66_RS22915 (LI66_22915) | 9799..10473 | - | 675 | WP_022649979 | prepilin peptidase | - |
| LI66_RS22920 (LI66_22920) | 10458..10940 | - | 483 | WP_045261878 | lytic transglycosylase domain-containing protein | virB1 |
| LI66_RS22925 (LI66_22925) | 11025..11639 | - | 615 | WP_022649977 | type 4 pilus major pilin | - |
| LI66_RS22930 (LI66_22930) | 11653..12747 | - | 1095 | WP_032623575 | type II secretion system F family protein | - |
| LI66_RS22935 (LI66_22935) | 12740..14299 | - | 1560 | WP_022649975 | GspE/PulE family protein | virB11 |
| LI66_RS22940 (LI66_22940) | 14302..14817 | - | 516 | WP_022649974 | type IV pilus biogenesis protein PilP | - |
| LI66_RS22945 (LI66_22945) | 14801..16105 | - | 1305 | WP_022649973 | type 4b pilus protein PilO2 | - |
| LI66_RS22950 (LI66_22950) | 16107..17783 | - | 1677 | WP_022649972 | PilN family type IVB pilus formation outer membrane protein | - |
| LI66_RS22955 (LI66_22955) | 17798..18235 | - | 438 | WP_022649971 | type IV pilus biogenesis protein PilM | - |
Region 2: 86553..103674
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LI66_RS25930 | 83569..83772 | - | 204 | WP_123839396 | hypothetical protein | - |
| LI66_RS23345 (LI66_23345) | 83906..84175 | - | 270 | WP_022650007 | hypothetical protein | - |
| LI66_RS23350 (LI66_23350) | 84249..86549 | - | 2301 | WP_022650006 | F-type conjugative transfer protein TrbC | - |
| LI66_RS24500 | 86553..87809 | - | 1257 | WP_126530067 | hypothetical protein | trbB |
| LI66_RS23360 (LI66_23360) | 87790..88998 | - | 1209 | WP_022650004 | hypothetical protein | trbA |
| LI66_RS23365 (LI66_23365) | 89319..89951 | - | 633 | WP_022650003 | plasmid IncI1-type surface exclusion protein ExcA | - |
| LI66_RS23370 (LI66_23370) | 90013..92172 | - | 2160 | WP_022650002 | DotA/TraY family protein | traY |
| LI66_RS23375 (LI66_23375) | 92230..92772 | - | 543 | WP_022650001 | conjugal transfer protein TraX | - |
| LI66_RS23380 (LI66_23380) | 92769..93971 | - | 1203 | WP_022650000 | conjugal transfer protein TraW | traW |
| LI66_RS26005 (LI66_23385) | 93984..94553 | - | 570 | WP_022649999 | hypothetical protein | traV |
| LI66_RS23390 (LI66_23390) | 94553..97606 | - | 3054 | WP_022649998 | hypothetical protein | traU |
| LI66_RS23395 (LI66_23395) | 97693..98418 | - | 726 | WP_063864836 | hypothetical protein | traT |
| LI66_RS23400 (LI66_23400) | 98602..99006 | - | 405 | WP_022649996 | DUF6750 family protein | traR |
| LI66_RS23405 (LI66_23405) | 99041..99565 | - | 525 | WP_022649995 | conjugal transfer protein TraQ | traQ |
| LI66_RS25935 (LI66_23410) | 99565..100338 | - | 774 | WP_123839398 | conjugal transfer protein TraP | traP |
| LI66_RS23415 (LI66_23415) | 100319..101626 | - | 1308 | WP_022649993 | conjugal transfer protein TraO | traO |
| LI66_RS23420 (LI66_23420) | 101626..102606 | - | 981 | WP_022649992 | DotH/IcmK family type IV secretion protein | traN |
| LI66_RS23425 (LI66_23425) | 102617..103234 | - | 618 | WP_179297403 | DotI/IcmL family type IV secretion protein | traM |
| LI66_RS23430 (LI66_23430) | 103324..103674 | - | 351 | WP_022649990 | hypothetical protein | traL |
Host bacterium
| ID | 21632 | GenBank | NZ_CP010385 |
| Plasmid name | p34399-106.698kb | Incompatibility group | - |
| Plasmid size | 106698 bp | Coordinate of oriT [Strand] | 80344..80430 [-] |
| Host baterium | Enterobacter hormaechei subsp. xiangfangensis strain 34399 |
Cargo genes
| Drug resistance gene | blaLAP-2, qnrS1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIC1, AcrIIA7 |