Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121017 |
| Name | oriT_pA2483imp-1 |
| Organism | Enterobacter hormaechei subsp. xiangfangensis strain A2483 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LC532225 (7553..7652 [+], 100 nt) |
| oriT length | 100 nt |
| IRs (inverted repeats) | 25..32, 35..42 (TAACCTGT..ACAGGTTA) |
| Location of nic site | 61..62 |
| Conserved sequence flanking the nic site |
TCCTGCCCGG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 100 nt
>oriT_pA2483imp-1
GAATAAGGGGAGAGTAAAGTTTGATAACCTGTTTACAGGTTAGCTAACTTTACATGTCCTGCCCGGCTTTCGCCGGTGTTTACGCCAAGGAAAGTCTACA
GAATAAGGGGAGAGTAAAGTTTGATAACCTGTTTACAGGTTAGCTAACTTTACATGTCCTGCCCGGCTTTCGCCGGTGTTTACGCCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file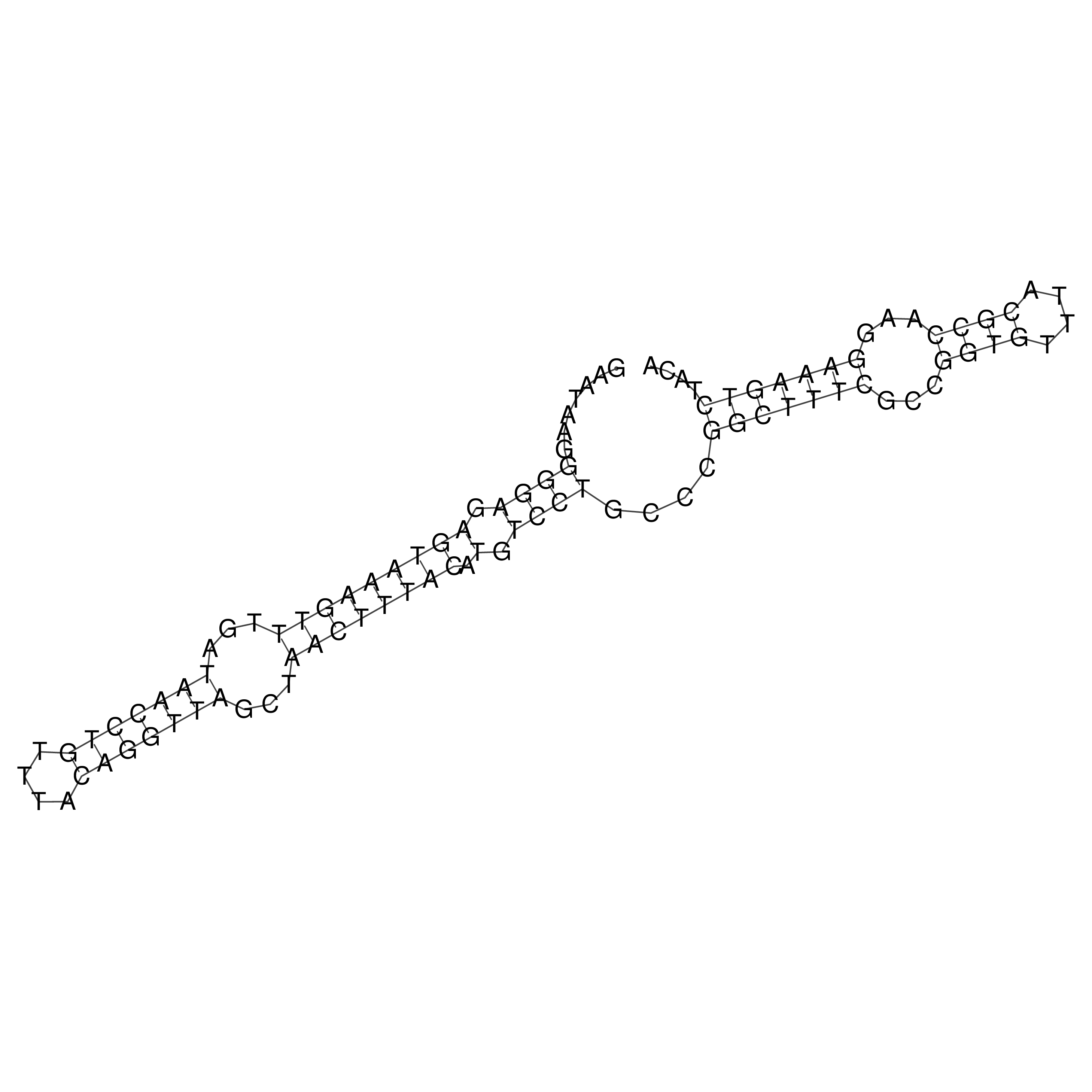
Relaxase
| ID | 13263 | GenBank | WP_077252989 |
| Name | traI_H5B13_RS00035_pA2483imp-1 |
UniProt ID | _ |
| Length | 870 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 870 a.a. Molecular weight: 96863.82 Da Isoelectric Point: 10.5262
>WP_077252989.1 TraI/MobA(P) family conjugative relaxase [Enterobacter hormaechei]
MRNVKKSSFVELAKYLQDPQSKQERVGVIKVTNCHQENILDAARYDIEPTQRRNTRSEGDKTYHLVISFR
PGEKPSQEVLDAAEERICEAMGYADHQRVSAVHYDTDNVHIHIAINKIHPQTHNIHTPYNDYKTRSAVCL
ALEKEFGLESDNHRASKTRSENNAADMERHSGRESLLGWIKRECADQLKSAANWQELHQAMRQNGLELRQ
KGNGFVITGPDGLAVKASSVDRSLSRAALEKKLGSFESSVPDTKPSRPSPEAIKQRFGSAAHVKRPGDRP
PPMRENRQVNLGDVPVMQINSGKRYEGKPLGSQGASTSALYGRYQSEQQRLTAARNEALRLAKVKRDRLI
ENAKRSGRLKRAAIKLLKGPGVNKRYLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWADWLQFKA
AGGDDEALKTLRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDGGKRLDVS
RDAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELNVSFADPELEKLRQILTKKAAREREY
IHIPFEQRGKARQAGAMWDTEKKSWFVGPYATRDRIEKFIRMNEEKDNEQRRQRYAGTERTDAGRERSGT
GNATGANEPGFTGRSTSADAGNGYGTSSSGRRTGTGDAFTTAGSQPGRSDDTRTGRSRAAEPNGERGHSK
PNVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLQGNVSDQLEHKTAERADQLRRDMAGAGGITTPA
EVKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIVVMPVSDY
TARRLSNLKVGESVTLTPSGSVRTTKGRSR
MRNVKKSSFVELAKYLQDPQSKQERVGVIKVTNCHQENILDAARYDIEPTQRRNTRSEGDKTYHLVISFR
PGEKPSQEVLDAAEERICEAMGYADHQRVSAVHYDTDNVHIHIAINKIHPQTHNIHTPYNDYKTRSAVCL
ALEKEFGLESDNHRASKTRSENNAADMERHSGRESLLGWIKRECADQLKSAANWQELHQAMRQNGLELRQ
KGNGFVITGPDGLAVKASSVDRSLSRAALEKKLGSFESSVPDTKPSRPSPEAIKQRFGSAAHVKRPGDRP
PPMRENRQVNLGDVPVMQINSGKRYEGKPLGSQGASTSALYGRYQSEQQRLTAARNEALRLAKVKRDRLI
ENAKRSGRLKRAAIKLLKGPGVNKRYLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWADWLQFKA
AGGDDEALKTLRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDGGKRLDVS
RDAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELNVSFADPELEKLRQILTKKAAREREY
IHIPFEQRGKARQAGAMWDTEKKSWFVGPYATRDRIEKFIRMNEEKDNEQRRQRYAGTERTDAGRERSGT
GNATGANEPGFTGRSTSADAGNGYGTSSSGRRTGTGDAFTTAGSQPGRSDDTRTGRSRAAEPNGERGHSK
PNVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLQGNVSDQLEHKTAERADQLRRDMAGAGGITTPA
EVKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIVVMPVSDY
TARRLSNLKVGESVTLTPSGSVRTTKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15545 | GenBank | WP_004196203 |
| Name | t4cp2_H5B13_RS00025_pA2483imp-1 |
UniProt ID | _ |
| Length | 648 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 648 a.a. Molecular weight: 71683.77 Da Isoelectric Point: 8.3842
>WP_004196203.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacterales]
MRKPNNAVGPQVRNKKKTARGKLIPAMAITSLAAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHDRLPDAFFAAGSVGATVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIKAAGLLGNDEGV
YVGAWEDKNGQLQYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYA
KNLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVL
YKHKNDGTPATLPYVDSIMADPERGAGELWMEMTQYGHVNGENHPVVGAAGRDMMDRPEEEAGSVLSTAK
SYLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERV
NNNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLAMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQD
INQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTM
QEVQRPLLTVDECLRMPGPKKNAEGLITERGDMVVYVAGFPAIYGKQPLFFQDEIFSLRASVPEPKTTDR
IRAAAAANDDASNEAIAI
MRKPNNAVGPQVRNKKKTARGKLIPAMAITSLAAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHDRLPDAFFAAGSVGATVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIKAAGLLGNDEGV
YVGAWEDKNGQLQYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYA
KNLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVL
YKHKNDGTPATLPYVDSIMADPERGAGELWMEMTQYGHVNGENHPVVGAAGRDMMDRPEEEAGSVLSTAK
SYLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERV
NNNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLAMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQD
INQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTM
QEVQRPLLTVDECLRMPGPKKNAEGLITERGDMVVYVAGFPAIYGKQPLFFQDEIFSLRASVPEPKTTDR
IRAAAAANDDASNEAIAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 27408..37651
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H5B13_RS00165 | 22520..23098 | - | 579 | WP_004196261 | hypothetical protein | - |
| H5B13_RS00170 | 23095..23343 | - | 249 | WP_004196202 | hypothetical protein | - |
| H5B13_RS00175 | 23343..23651 | - | 309 | WP_003019666 | HigA family addiction module antitoxin | - |
| H5B13_RS00180 | 23660..23959 | - | 300 | WP_003019663 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| H5B13_RS00185 | 25286..26179 | - | 894 | WP_004196197 | plasmid replication initiator TrfA | - |
| H5B13_RS00190 | 26273..26644 | - | 372 | WP_181708861 | single-stranded DNA-binding protein | - |
| H5B13_RS00195 | 26750..27154 | + | 405 | WP_003019654 | transcriptional regulator | - |
| H5B13_RS00200 | 27408..28295 | + | 888 | WP_003019652 | P-type conjugative transfer ATPase TrbB | virB11 |
| H5B13_RS00205 | 28409..28792 | + | 384 | WP_003019650 | TrbC/VirB2 family protein | virB2 |
| H5B13_RS00210 | 28795..29121 | + | 327 | WP_003019648 | conjugal transfer protein TrbD | virB3 |
| H5B13_RS00215 | 29118..31661 | + | 2544 | WP_004196242 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| H5B13_RS00220 | 31658..32365 | + | 708 | WP_004196182 | VirB8/TrbF family protein | virB8 |
| H5B13_RS00225 | 32381..33271 | + | 891 | WP_003019641 | P-type conjugative transfer protein TrbG | virB9 |
| H5B13_RS00230 | 33274..33726 | + | 453 | WP_003019638 | conjugal transfer protein TrbH | - |
| H5B13_RS00235 | 33730..35124 | + | 1395 | WP_003019636 | TrbI/VirB10 family protein | virB10 |
| H5B13_RS00240 | 35137..35907 | + | 771 | WP_004196177 | P-type conjugative transfer protein TrbJ | virB5 |
| H5B13_RS00245 | 35920..36093 | + | 174 | WP_004196247 | entry exclusion lipoprotein TrbK | - |
| H5B13_RS00250 | 36098..37651 | + | 1554 | WP_023363220 | P-type conjugative transfer protein TrbL | virB6 |
| H5B13_RS00255 | 37662..37967 | + | 306 | WP_003019624 | IncN plasmid KikA protein | - |
| H5B13_RS00260 | 37983..38753 | + | 771 | WP_004196237 | transglycosylase SLT domain protein | - |
| H5B13_RS00265 | 38770..39456 | + | 687 | WP_004196226 | TraX family protein | - |
| H5B13_RS00435 | 39902..39997 | + | 96 | WP_003019619 | DinQ-like type I toxin DqlB | - |
| H5B13_RS00270 | 40058..40378 | - | 321 | WP_004196195 | hypothetical protein | - |
| H5B13_RS00275 | 40961..41191 | - | 231 | WP_004196204 | hypothetical protein | - |
| H5B13_RS00280 | 41344..41847 | - | 504 | WP_004196173 | thermonuclease family protein | - |
| H5B13_RS00285 | 41847..42101 | - | 255 | WP_004196245 | hypothetical protein | - |
| H5B13_RS00290 | 42091..42414 | - | 324 | WP_004196208 | hypothetical protein | - |
Host bacterium
| ID | 21446 | GenBank | NZ_LC532225 |
| Plasmid name | pA2483imp-1 | Incompatibility group | - |
| Plasmid size | 61594 bp | Coordinate of oriT [Strand] | 7553..7652 [+] |
| Host baterium | Enterobacter hormaechei subsp. xiangfangensis strain A2483 |
Cargo genes
| Drug resistance gene | sul1, qacE, aac(6')-IIc, blaIMP-1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |