Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121001 |
| Name | oriT_pA2504imp-1 |
| Organism | Enterobacter hormaechei subsp. xiangfangensis strain A2504 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LC532227 (28639..28738 [+], 100 nt) |
| oriT length | 100 nt |
| IRs (inverted repeats) | 25..32, 35..42 (TAACCTGT..ACAGGTTA) |
| Location of nic site | 61..62 |
| Conserved sequence flanking the nic site |
TCCTGCCCGG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 100 nt
>oriT_pA2504imp-1
GAATAAGGGGAGAGTAAAGTTTGATAACCTGTTTACAGGTTAGCTAACTTTACATGTCCTGCCCGGCTTTCGCCGGTGTTTACGCCAAGGAAAGTCTACA
GAATAAGGGGAGAGTAAAGTTTGATAACCTGTTTACAGGTTAGCTAACTTTACATGTCCTGCCCGGCTTTCGCCGGTGTTTACGCCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file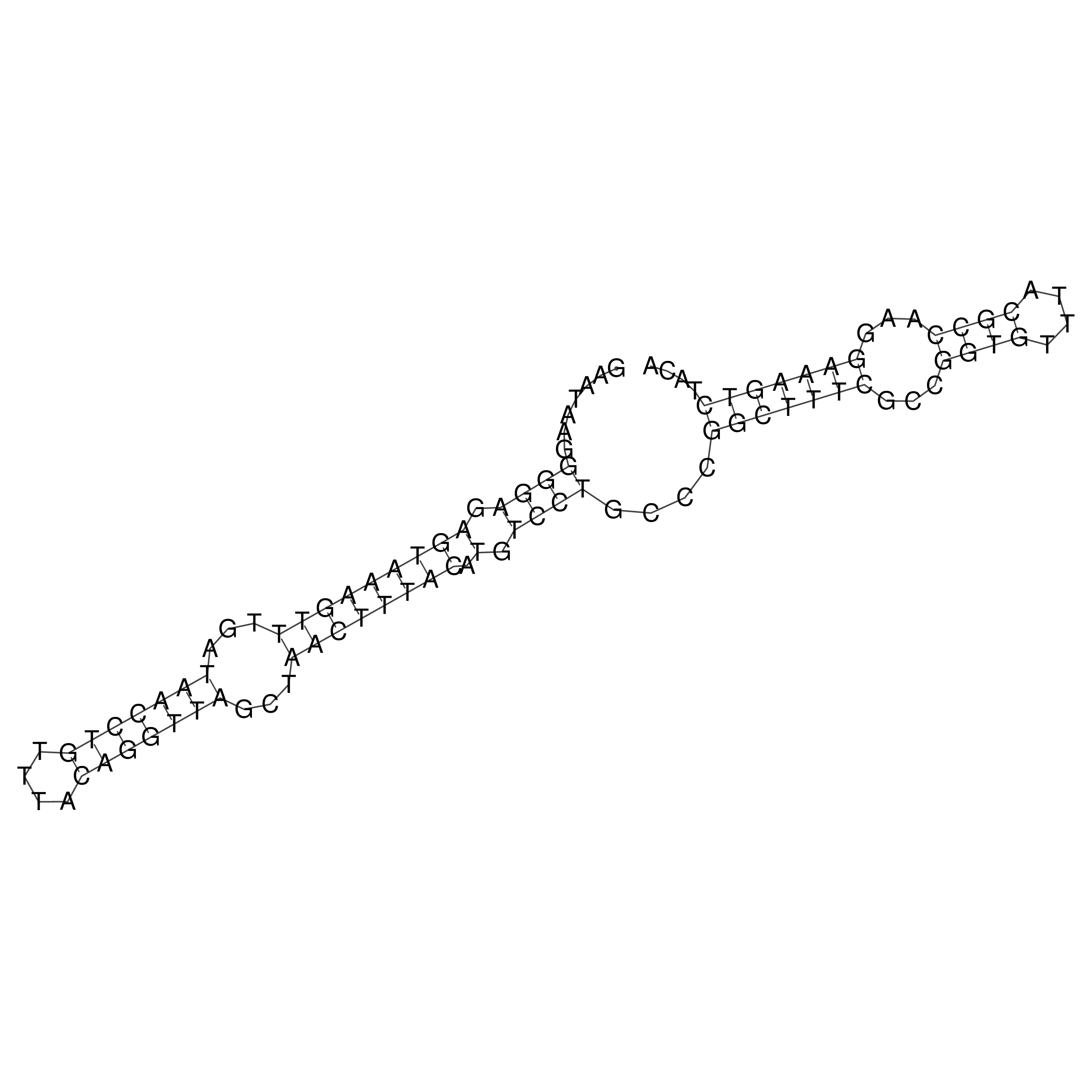
Relaxase
| ID | 13250 | GenBank | WP_004196169 |
| Name | traI_H5B15_RS00185_pA2504imp-1 |
UniProt ID | _ |
| Length | 877 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 877 a.a. Molecular weight: 97698.86 Da Isoelectric Point: 10.5465
>WP_004196169.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Enterobacteriaceae]
MISRHIPMRNVKKSSFVELAKYLQDPQSKQERVGVIKVTNCHQENILDAARYDIEPTQRRNTRSEGDKTY
HLVISFRPGEKPSQEVLDAAEERICEAMGYADHQRVSAVHYDTDNVHIHIAINKIHPQTHNIHTPYNDYK
TRSAVCLALEKEFGLESDNHRASKTRSENNAADMERHSGRESLLGWIKRECADQLKSAANWQELHQAMRQ
NGLELRQKGNGFVITGPDGLAVKASSVDRSLSRAALEKKLGSFESSVPDTKPSRPSPEAIKQRFGSAAHV
KRPGDRPPPMRENRQVNLGDVPVMQINSGKRYEGKPLGSQGASTSALYGRYQSEQQRLTAARNEALRLAK
VKRDRLIENAKRSGRLKRAAIKLLKGPGVNKRYLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWA
DWLQFKAAGGDDEALKTLRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDG
GKRLDVSRDAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELNVSFADPELEKLRQILTKK
AAREREYIHIPFEQRGKARQAGAMWDTEKKSWFVGPYATRDRIEKFIRMNEEKDNEQRRQRYAGTERTDA
GRERSGTGNATGANEPGFTGRSTSADAGNGYGTSSSGRRTGTGDAFTTAGSQPGRSDDTRTGRSRAAEPN
GERGHSKPNVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLQGNVSDQLEHKTAERADQLRRDMAGA
GGITTPAEVKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIV
VMPVSDYTARRLSNLKVGESVTLTPSGSVRTTKGRSR
MISRHIPMRNVKKSSFVELAKYLQDPQSKQERVGVIKVTNCHQENILDAARYDIEPTQRRNTRSEGDKTY
HLVISFRPGEKPSQEVLDAAEERICEAMGYADHQRVSAVHYDTDNVHIHIAINKIHPQTHNIHTPYNDYK
TRSAVCLALEKEFGLESDNHRASKTRSENNAADMERHSGRESLLGWIKRECADQLKSAANWQELHQAMRQ
NGLELRQKGNGFVITGPDGLAVKASSVDRSLSRAALEKKLGSFESSVPDTKPSRPSPEAIKQRFGSAAHV
KRPGDRPPPMRENRQVNLGDVPVMQINSGKRYEGKPLGSQGASTSALYGRYQSEQQRLTAARNEALRLAK
VKRDRLIENAKRSGRLKRAAIKLLKGPGVNKRYLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWA
DWLQFKAAGGDDEALKTLRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDG
GKRLDVSRDAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELNVSFADPELEKLRQILTKK
AAREREYIHIPFEQRGKARQAGAMWDTEKKSWFVGPYATRDRIEKFIRMNEEKDNEQRRQRYAGTERTDA
GRERSGTGNATGANEPGFTGRSTSADAGNGYGTSSSGRRTGTGDAFTTAGSQPGRSDDTRTGRSRAAEPN
GERGHSKPNVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLQGNVSDQLEHKTAERADQLRRDMAGA
GGITTPAEVKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIV
VMPVSDYTARRLSNLKVGESVTLTPSGSVRTTKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15529 | GenBank | WP_004196203 |
| Name | t4cp2_H5B15_RS00175_pA2504imp-1 |
UniProt ID | _ |
| Length | 648 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 648 a.a. Molecular weight: 71683.77 Da Isoelectric Point: 8.3842
>WP_004196203.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacterales]
MRKPNNAVGPQVRNKKKTARGKLIPAMAITSLAAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHDRLPDAFFAAGSVGATVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIKAAGLLGNDEGV
YVGAWEDKNGQLQYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYA
KNLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVL
YKHKNDGTPATLPYVDSIMADPERGAGELWMEMTQYGHVNGENHPVVGAAGRDMMDRPEEEAGSVLSTAK
SYLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERV
NNNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLAMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQD
INQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTM
QEVQRPLLTVDECLRMPGPKKNAEGLITERGDMVVYVAGFPAIYGKQPLFFQDEIFSLRASVPEPKTTDR
IRAAAAANDDASNEAIAI
MRKPNNAVGPQVRNKKKTARGKLIPAMAITSLAAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHDRLPDAFFAAGSVGATVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIKAAGLLGNDEGV
YVGAWEDKNGQLQYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYA
KNLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVL
YKHKNDGTPATLPYVDSIMADPERGAGELWMEMTQYGHVNGENHPVVGAAGRDMMDRPEEEAGSVLSTAK
SYLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERV
NNNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLAMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQD
INQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTM
QEVQRPLLTVDECLRMPGPKKNAEGLITERGDMVVYVAGFPAIYGKQPLFFQDEIFSLRASVPEPKTTDR
IRAAAAANDDASNEAIAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 48425..58737
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H5B15_RS00315 | 43606..44184 | - | 579 | WP_004196261 | hypothetical protein | - |
| H5B15_RS00320 | 44181..44429 | - | 249 | WP_004196202 | hypothetical protein | - |
| H5B15_RS00325 | 44429..44737 | - | 309 | WP_003019666 | HigA family addiction module antitoxin | - |
| H5B15_RS00330 | 44746..45045 | - | 300 | WP_003019663 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| H5B15_RS00335 | 46372..47265 | - | 894 | WP_004196197 | plasmid replication initiator TrfA | - |
| H5B15_RS00340 | 47359..47730 | - | 372 | WP_181708861 | single-stranded DNA-binding protein | - |
| H5B15_RS00345 | 47836..48240 | + | 405 | WP_003019654 | transcriptional regulator | - |
| H5B15_RS00350 | 48425..49381 | + | 957 | WP_032932562 | P-type conjugative transfer ATPase TrbB | virB11 |
| H5B15_RS00355 | 49495..49878 | + | 384 | WP_003019650 | TrbC/VirB2 family protein | virB2 |
| H5B15_RS00360 | 49881..50207 | + | 327 | WP_003019648 | conjugal transfer protein TrbD | virB3 |
| H5B15_RS00365 | 50204..52747 | + | 2544 | WP_004196242 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| H5B15_RS00370 | 52744..53451 | + | 708 | WP_004196182 | VirB8/TrbF family protein | virB8 |
| H5B15_RS00375 | 53467..54357 | + | 891 | WP_003019641 | P-type conjugative transfer protein TrbG | virB9 |
| H5B15_RS00380 | 54360..54812 | + | 453 | WP_003019638 | conjugal transfer protein TrbH | - |
| H5B15_RS00385 | 54816..56210 | + | 1395 | WP_003019636 | TrbI/VirB10 family protein | virB10 |
| H5B15_RS00390 | 56223..56993 | + | 771 | WP_004196177 | P-type conjugative transfer protein TrbJ | virB5 |
| H5B15_RS00395 | 57006..57179 | + | 174 | WP_004196247 | entry exclusion lipoprotein TrbK | - |
| H5B15_RS00400 | 57184..58737 | + | 1554 | WP_023363220 | P-type conjugative transfer protein TrbL | virB6 |
| H5B15_RS00405 | 58748..59053 | + | 306 | WP_003019624 | IncN plasmid KikA protein | - |
| H5B15_RS00410 | 59069..59839 | + | 771 | WP_004196237 | transglycosylase SLT domain protein | - |
| H5B15_RS00415 | 59856..60542 | + | 687 | WP_004196226 | TraX family protein | - |
| H5B15_RS00435 | 60988..61083 | + | 96 | WP_003019619 | DinQ-like type I toxin DqlB | - |
| H5B15_RS00420 | 61144..61464 | - | 321 | WP_004196195 | hypothetical protein | - |
Host bacterium
| ID | 21430 | GenBank | NZ_LC532227 |
| Plasmid name | pA2504imp-1 | Incompatibility group | - |
| Plasmid size | 61594 bp | Coordinate of oriT [Strand] | 28639..28738 [+] |
| Host baterium | Enterobacter hormaechei subsp. xiangfangensis strain A2504 |
Cargo genes
| Drug resistance gene | sul1, qacE, aac(6')-IIc, blaIMP-1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |