Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120995 |
| Name | oriT_pW2_73_2 |
| Organism | Agrobacterium rubi strain W2/73 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP049209 (98495..98537 [-], 43 nt) |
| oriT length | 43 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 43 nt
>oriT_pW2_73_2
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCGTTGC
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCGTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file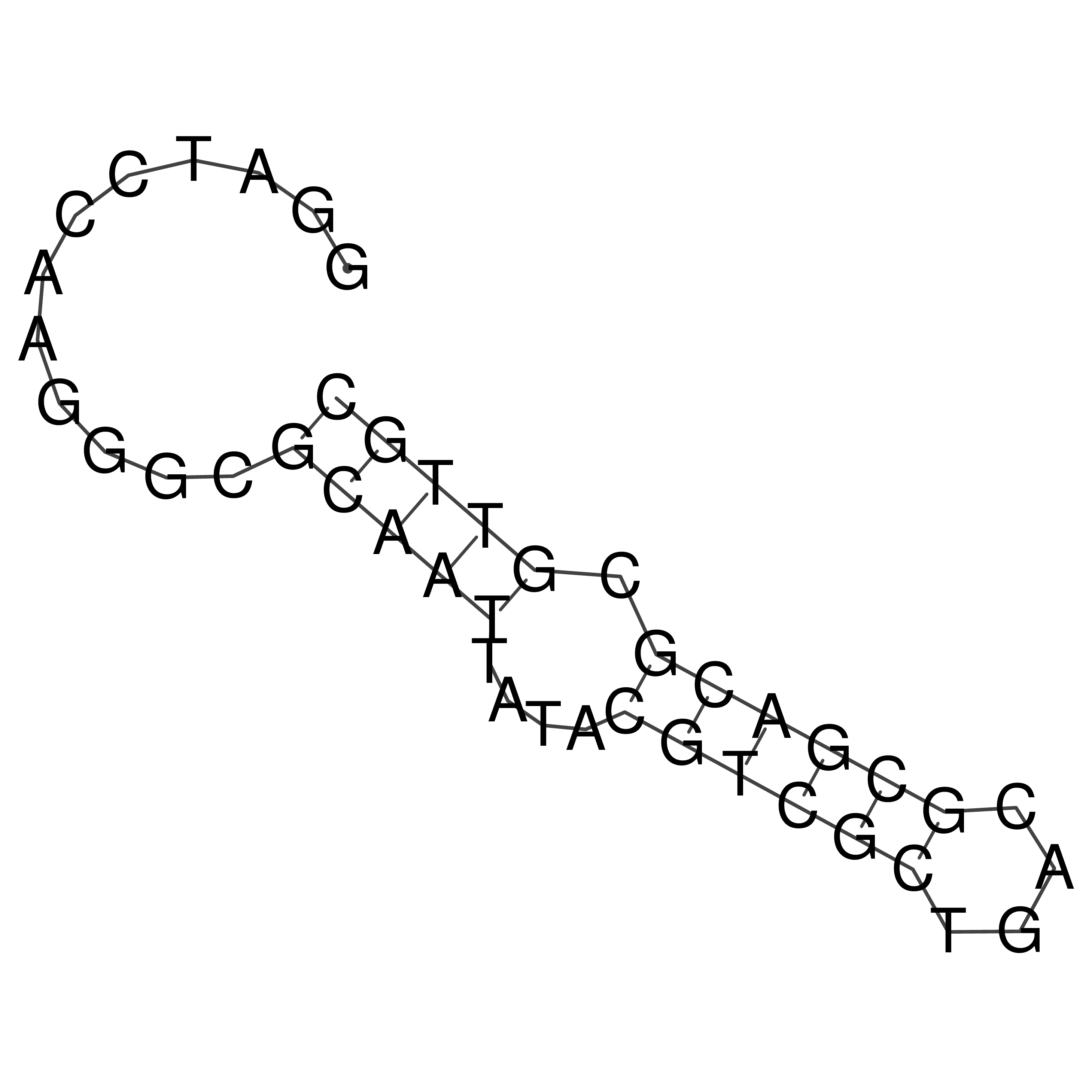
Relaxase
| ID | 13245 | GenBank | WP_065699131 |
| Name | traA_G6M88_RS24275_pW2_73_2 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123869.00 Da Isoelectric Point: 10.0632
>WP_065699131.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium rubi]
MAIAHFSASIVSRGRGASVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPADAPKWVRAMIADR
SVSGASEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHIHAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEEGFGSKKVAVIGEDRQPVRTKSGKILYQLWAGSTDDFSVLRDGWFERLNHHLALGG
IDLKIDGRSYEKQGIELEPTIHLGVGTKAIERKAQEQGVRPELERIELNEELRSENTRRILRNPAIVLDL
ITREKSVFDERDIAKVLHRYVDDPAVFLQLMARIIQNPEALRLQRDTIKFATGEKVPARYSTIAMIRLEA
TMVRQALWLSHRNSRAVSEAALAATFRRHERLSEEQRTAIERIAGPARIAAVVGRAGAGKTTMMKAAREV
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRTIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
VNTRIKGTRLKAEAVERLIGDWNNDYDQTKTTLILAHLRRDVRMLNVMAREKLVERGIVGEGYVFKTADG
VRRFDVGDQIVFLKNETSLGVKNGMIAHVIEAAPNKIVAVVGEGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFSGGLAKVLSRRQAKETTLDYERGKLY
REALRFAESRGLNIVQVARTLIRDQLDWTLRQKMKLVDLGQRLAVFAERLGLHQPPMTQTMKEAAPMVAG
IKTFSGSVADTVEHKLDTDPALRQQWEEVSTRFAYVFADPETAFRAMNFDAVLAHKETAKLALQTLTTAP
ASIGPLRGKTGILASKVEREARRIAEINAPALKRDLEQYLQMREAATKRLQADEQALRQRVSIDIPALSP
AARVVLERVRDAIDRNDVPAAMAYALSNRETKLEIDGFNKAITERFGERTLLTNAAREPSGKLFDKLSDG
MQPQQKERLKEAWPIMRTSQQLAAHERTIQTLRQAEDLRLAQRQTPVMKQ
MAIAHFSASIVSRGRGASVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPADAPKWVRAMIADR
SVSGASEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHIHAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEEGFGSKKVAVIGEDRQPVRTKSGKILYQLWAGSTDDFSVLRDGWFERLNHHLALGG
IDLKIDGRSYEKQGIELEPTIHLGVGTKAIERKAQEQGVRPELERIELNEELRSENTRRILRNPAIVLDL
ITREKSVFDERDIAKVLHRYVDDPAVFLQLMARIIQNPEALRLQRDTIKFATGEKVPARYSTIAMIRLEA
TMVRQALWLSHRNSRAVSEAALAATFRRHERLSEEQRTAIERIAGPARIAAVVGRAGAGKTTMMKAAREV
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRTIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
VNTRIKGTRLKAEAVERLIGDWNNDYDQTKTTLILAHLRRDVRMLNVMAREKLVERGIVGEGYVFKTADG
VRRFDVGDQIVFLKNETSLGVKNGMIAHVIEAAPNKIVAVVGEGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFSGGLAKVLSRRQAKETTLDYERGKLY
REALRFAESRGLNIVQVARTLIRDQLDWTLRQKMKLVDLGQRLAVFAERLGLHQPPMTQTMKEAAPMVAG
IKTFSGSVADTVEHKLDTDPALRQQWEEVSTRFAYVFADPETAFRAMNFDAVLAHKETAKLALQTLTTAP
ASIGPLRGKTGILASKVEREARRIAEINAPALKRDLEQYLQMREAATKRLQADEQALRQRVSIDIPALSP
AARVVLERVRDAIDRNDVPAAMAYALSNRETKLEIDGFNKAITERFGERTLLTNAAREPSGKLFDKLSDG
MQPQQKERLKEAWPIMRTSQQLAAHERTIQTLRQAEDLRLAQRQTPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15526 | GenBank | WP_065699101 |
| Name | t4cp2_G6M88_RS24080_pW2_73_2 |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 92138.01 Da Isoelectric Point: 5.8903
>WP_065699101.1 conjugal transfer protein TrbE [Agrobacterium rubi]
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVETIRVATGDYPTETEAHFPDPVTRAIDEERRMHFQKEKGHFESRHALILTWRPPEPRRSRLTR
YIYSDAASRSATYADKALTSFLTSIREVEQYLANVVSIRRMVTRETSERGGYRVARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPLVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAISEASSQLVAYGYYTPVIVI
FEEDPTRLQEKSEAIRRLVQAEGFGARIETLNATDAFLGSLPGISYANIREPLINTRNLADLIPLNSVWS
GSPNAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGSSMLPLTLGLNGDHYQIGGDIGSSASGEVRALSFCPLAELSSDGDRAWAAEWIEMLVALQGVTITP
DYRNAISRQVALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGPFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGDPSLIILDEAWLMLGHPVFRDKIRDWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNEQQIEIVATAIPKREYYVVSPEGRRLFDM
ALGPVALSFVGATGKKDLKRIRILHSEHGDAWPLHWLQQRGIAHAETLFPTF
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVETIRVATGDYPTETEAHFPDPVTRAIDEERRMHFQKEKGHFESRHALILTWRPPEPRRSRLTR
YIYSDAASRSATYADKALTSFLTSIREVEQYLANVVSIRRMVTRETSERGGYRVARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPLVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAISEASSQLVAYGYYTPVIVI
FEEDPTRLQEKSEAIRRLVQAEGFGARIETLNATDAFLGSLPGISYANIREPLINTRNLADLIPLNSVWS
GSPNAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGSSMLPLTLGLNGDHYQIGGDIGSSASGEVRALSFCPLAELSSDGDRAWAAEWIEMLVALQGVTITP
DYRNAISRQVALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGPFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGDPSLIILDEAWLMLGHPVFRDKIRDWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNEQQIEIVATAIPKREYYVVSPEGRRLFDM
ALGPVALSFVGATGKKDLKRIRILHSEHGDAWPLHWLQQRGIAHAETLFPTF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 50992..60684
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| G6M88_RS26210 | 46132..46287 | - | 156 | WP_234791161 | hypothetical protein | - |
| G6M88_RS24045 (G6M88_24070) | 46347..47561 | - | 1215 | WP_065699094 | plasmid replication protein RepC | - |
| G6M88_RS24050 (G6M88_24075) | 47735..48694 | - | 960 | WP_065699095 | plasmid partitioning protein RepB | - |
| G6M88_RS24055 (G6M88_24080) | 48797..49993 | - | 1197 | WP_065699096 | plasmid partitioning protein RepA | - |
| G6M88_RS24060 (G6M88_24085) | 50360..50995 | + | 636 | WP_065699097 | acyl-homoserine-lactone synthase TraI | - |
| G6M88_RS24065 (G6M88_24090) | 50992..51963 | + | 972 | WP_065699098 | P-type conjugative transfer ATPase TrbB | virB11 |
| G6M88_RS24070 (G6M88_24095) | 51953..52357 | + | 405 | WP_065699099 | conjugal transfer pilin TrbC | virB2 |
| G6M88_RS24075 (G6M88_24100) | 52350..52649 | + | 300 | WP_065699100 | conjugal transfer protein TrbD | virB3 |
| G6M88_RS24080 (G6M88_24105) | 52659..55127 | + | 2469 | WP_065699101 | conjugal transfer protein TrbE | virb4 |
| G6M88_RS24085 (G6M88_24110) | 55099..55908 | + | 810 | WP_065699102 | P-type conjugative transfer protein TrbJ | virB5 |
| G6M88_RS24090 (G6M88_24115) | 55905..56129 | + | 225 | WP_065699103 | entry exclusion protein TrbK | - |
| G6M88_RS24095 (G6M88_24120) | 56123..57313 | + | 1191 | WP_065699104 | P-type conjugative transfer protein TrbL | virB6 |
| G6M88_RS24100 (G6M88_24125) | 57331..58008 | + | 678 | WP_065699105 | conjugal transfer protein TrbF | virB8 |
| G6M88_RS24105 (G6M88_24130) | 58026..58877 | + | 852 | WP_045232178 | P-type conjugative transfer protein TrbG | virB9 |
| G6M88_RS24110 (G6M88_24135) | 58877..59353 | + | 477 | WP_065699106 | conjugal transfer protein TrbH | - |
| G6M88_RS24115 (G6M88_24140) | 59365..60684 | + | 1320 | WP_065699107 | IncP-type conjugal transfer protein TrbI | virB10 |
| G6M88_RS24120 (G6M88_24145) | 60944..63439 | - | 2496 | WP_234791162 | HAMP domain-containing methyl-accepting chemotaxis protein | - |
| G6M88_RS24125 (G6M88_24150) | 64358..65560 | - | 1203 | WP_065699109 | GTP-binding protein | - |
Host bacterium
| ID | 21424 | GenBank | NZ_CP049209 |
| Plasmid name | pW2_73_2 | Incompatibility group | - |
| Plasmid size | 179725 bp | Coordinate of oriT [Strand] | 98495..98537 [-] |
| Host baterium | Agrobacterium rubi strain W2/73 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsH |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |