Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120994 |
| Name | oriT_pW2_73_1 |
| Organism | Agrobacterium rubi strain W2/73 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP049208 (44043..44076 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pW2_73_1
TCCAAGGGCGCAATTATACGTCGCAGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCAGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file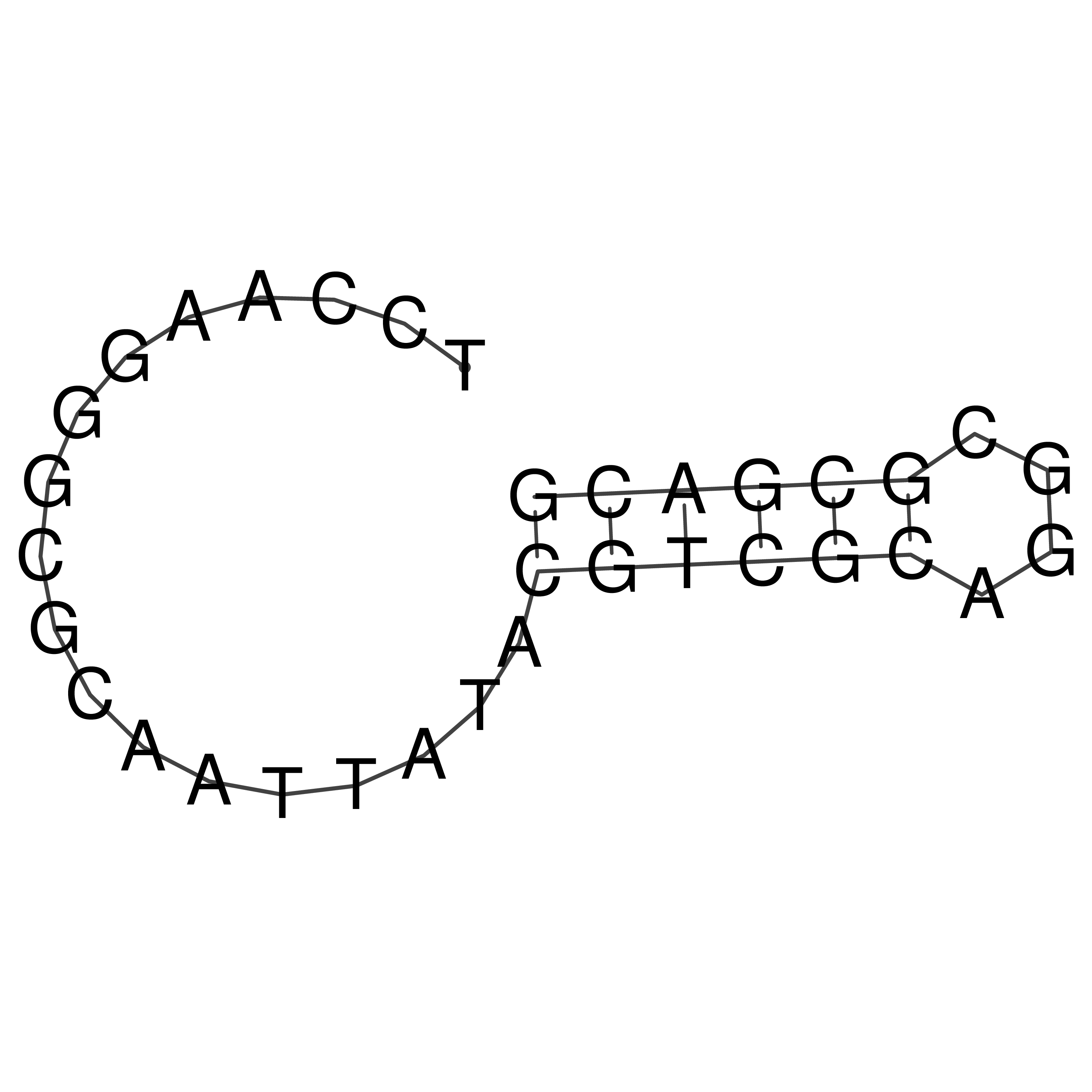
Relaxase
| ID | 13244 | GenBank | WP_065700853 |
| Name | traA_G6M88_RS22705_pW2_73_1 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123249.06 Da Isoelectric Point: 9.7724
>WP_065700853.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium rubi]
MAVPHFSVSIVARSSGRSAVLSAAYRHCARMEFEREARTIDYTRKQGLLHEAFVIPDDAPEWVRSMMADR
SVAGASEAFWNKVEDFEKRSDAQLAKDVTIALPLELTAEQNIELMRDFVTSHITSKGMVADWVYHDAPGN
PHVHLMATLRPLTEDGFGSKKVAVTGPEGKPIRNDAGKIVYELWAGSAEDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELEPTIHLGVGTKAIARKAEETNRKQQRPASKLERIEIQEQRRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRVLQSPEALRLERERINFATGIRSPAKYTTREM
IRLEAEMAHRAIWLSGSPSHGVPEAVLQATFARHSRLSEEQRTAISHVAAPARIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGSLAGKAAEGLDKDAGIVSRTLSSWELRWSQGRNQLDDKTVFVLDEAGMVSSRQM
ALLVETVTKAGAKLVLVGDPQQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVGQA
VDAYRAHGYMRGSDLKAQAVDKLITDWDRDFDPSRTSLILAHLRRDVRKLNDMARARLVDRGVLEDGFAF
KTEDGSRMFATGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEVGDGEHRRQVTVEQRFYNTLDHGYA
TTIHKAQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFSKAGGLTQILSRQNAKETSLDYE
QSTLYRHALRFAEARGLHLMNVARTIAQDRLQWAVRQKHKLADLGTRLAAIGTALGLALGSNKHAITDTI
NEARPMVSGITIFPKSLDQAVEDKLAADPGLKMQWDDVSTRFHFVYANPQAAFKAIDVDTMLKDPSVAQF
TLAKIAHQPESFGTLKGKTGLLASRGDRQDREKAIANVPALARNLERYLRERAEAEIKHETEERAVRQKV
SVDIPALSPSAKQTLERVRDAIDRNDLSAGLEYALADKMVKAELEGFARAVSERFGERTFLPLAAKDTGG
KTFETVTSGMTVGQKAEVQSAWNSMRTVQQLGAHERTTEAFRQAETMRQTKSQGLSLK
MAVPHFSVSIVARSSGRSAVLSAAYRHCARMEFEREARTIDYTRKQGLLHEAFVIPDDAPEWVRSMMADR
SVAGASEAFWNKVEDFEKRSDAQLAKDVTIALPLELTAEQNIELMRDFVTSHITSKGMVADWVYHDAPGN
PHVHLMATLRPLTEDGFGSKKVAVTGPEGKPIRNDAGKIVYELWAGSAEDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELEPTIHLGVGTKAIARKAEETNRKQQRPASKLERIEIQEQRRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRVLQSPEALRLERERINFATGIRSPAKYTTREM
IRLEAEMAHRAIWLSGSPSHGVPEAVLQATFARHSRLSEEQRTAISHVAAPARIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGSLAGKAAEGLDKDAGIVSRTLSSWELRWSQGRNQLDDKTVFVLDEAGMVSSRQM
ALLVETVTKAGAKLVLVGDPQQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVGQA
VDAYRAHGYMRGSDLKAQAVDKLITDWDRDFDPSRTSLILAHLRRDVRKLNDMARARLVDRGVLEDGFAF
KTEDGSRMFATGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEVGDGEHRRQVTVEQRFYNTLDHGYA
TTIHKAQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFSKAGGLTQILSRQNAKETSLDYE
QSTLYRHALRFAEARGLHLMNVARTIAQDRLQWAVRQKHKLADLGTRLAAIGTALGLALGSNKHAITDTI
NEARPMVSGITIFPKSLDQAVEDKLAADPGLKMQWDDVSTRFHFVYANPQAAFKAIDVDTMLKDPSVAQF
TLAKIAHQPESFGTLKGKTGLLASRGDRQDREKAIANVPALARNLERYLRERAEAEIKHETEERAVRQKV
SVDIPALSPSAKQTLERVRDAIDRNDLSAGLEYALADKMVKAELEGFARAVSERFGERTFLPLAAKDTGG
KTFETVTSGMTVGQKAEVQSAWNSMRTVQQLGAHERTTEAFRQAETMRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15524 | GenBank | WP_065700841 |
| Name | t4cp2_G6M88_RS22630_pW2_73_1 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91867.65 Da Isoelectric Point: 5.6923
>WP_065700841.1 MULTISPECIES: conjugal transfer protein TrbE [Agrobacterium]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLELNELSRQINAVLSRLG
SGWMIQVEAIRVPTVDYPSEDRCHFPDPVTRAIDAERRAHFEHEQGHFESKHAIVLTYRPLASKRTSLSK
YIYSDEESRKKSYADTVLFVFKSAIREVEQYFANTLSIRRMETRETIERGGERIARYDELLQFAKFCITG
ESHPVRLPDVPMYLDWIATAEFEHGLTPKVENRFLGIVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVAETEAAIAQASSKLVAYGYYTPVVVL
FDSDHEALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPVSPSPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLATGGDHYELGGDIAEEGRDLAFCPLSELQTDADRAWATEWIEMLVGLQGITITPDHRN
AISRQVGLMAKASGRSLSDFVSGVQLREVKDALHHYTVDGPMGQLLDAEEDGLTLGTFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARETGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGATGKEDLKRVRILKSEHGREWPIRWLETRGVHDAASLLKYE
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLELNELSRQINAVLSRLG
SGWMIQVEAIRVPTVDYPSEDRCHFPDPVTRAIDAERRAHFEHEQGHFESKHAIVLTYRPLASKRTSLSK
YIYSDEESRKKSYADTVLFVFKSAIREVEQYFANTLSIRRMETRETIERGGERIARYDELLQFAKFCITG
ESHPVRLPDVPMYLDWIATAEFEHGLTPKVENRFLGIVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVAETEAAIAQASSKLVAYGYYTPVVVL
FDSDHEALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPVSPSPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLATGGDHYELGGDIAEEGRDLAFCPLSELQTDADRAWATEWIEMLVGLQGITITPDHRN
AISRQVGLMAKASGRSLSDFVSGVQLREVKDALHHYTVDGPMGQLLDAEEDGLTLGTFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARETGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGATGKEDLKRVRILKSEHGREWPIRWLETRGVHDAASLLKYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15525 | GenBank | WP_065700856 |
| Name | traG_G6M88_RS22720_pW2_73_1 |
UniProt ID | _ |
| Length | 646 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 646 a.a. Molecular weight: 69454.52 Da Isoelectric Point: 9.1262
>WP_065700856.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Agrobacterium]
MAINRIALAVVPGTLMIVVVIAMTGIEQWLSGFGKTGSARLAWGRAGIALPYVSSAAIGILFLFASAGSI
NIRQAGCGVLAGCSGAILVAAARETMRLAAFTETLDADMSVLAYLDPATAIGAGVALMCACFALRVVLMG
NAAFASAEPKRIQGKRALHGKADWMKLTEAANLFPDAGGIVIGERYRVDKDSVGARAFRAGSAETWGVGG
KSPLLCFDGAFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGRAGRDVFVLD
PNTPDVGFNALDWVGRFGGTKEEDIASVASWIMSDSSGARGVRDDFFRASALQLLTALIADVCLSGHTDE
KNQTLRQVRMNLSEPEPTLRKRLQNIYDNSDSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGSSDVFININLKTLEAHSGLARVIIGSFLNAIYNRDGQMKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAGINDPETADYISRRCGVTT
VEIDQISLSWQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGHAPLRCGRAIWFRRDDMRACVD
GNRFHSNPSVPPTKTK
MAINRIALAVVPGTLMIVVVIAMTGIEQWLSGFGKTGSARLAWGRAGIALPYVSSAAIGILFLFASAGSI
NIRQAGCGVLAGCSGAILVAAARETMRLAAFTETLDADMSVLAYLDPATAIGAGVALMCACFALRVVLMG
NAAFASAEPKRIQGKRALHGKADWMKLTEAANLFPDAGGIVIGERYRVDKDSVGARAFRAGSAETWGVGG
KSPLLCFDGAFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGRAGRDVFVLD
PNTPDVGFNALDWVGRFGGTKEEDIASVASWIMSDSSGARGVRDDFFRASALQLLTALIADVCLSGHTDE
KNQTLRQVRMNLSEPEPTLRKRLQNIYDNSDSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGSSDVFININLKTLEAHSGLARVIIGSFLNAIYNRDGQMKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAGINDPETADYISRRCGVTT
VEIDQISLSWQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGHAPLRCGRAIWFRRDDMRACVD
GNRFHSNPSVPPTKTK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 25197..38902
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| G6M88_RS22595 (G6M88_22605) | 20506..21750 | - | 1245 | WP_065700834 | plasmid replication protein RepC | - |
| G6M88_RS22600 (G6M88_22610) | 21906..22901 | - | 996 | WP_065700835 | plasmid partitioning protein RepB | - |
| G6M88_RS22605 (G6M88_22615) | 22986..24176 | - | 1191 | WP_065700836 | plasmid partitioning protein RepA | - |
| G6M88_RS22610 (G6M88_22620) | 24562..25200 | + | 639 | WP_065700837 | acyl-homoserine-lactone synthase | - |
| G6M88_RS22615 (G6M88_22625) | 25197..26174 | + | 978 | WP_065700838 | P-type conjugative transfer ATPase TrbB | virB11 |
| G6M88_RS22620 (G6M88_22630) | 26164..26547 | + | 384 | WP_065700839 | TrbC/VirB2 family protein | virB2 |
| G6M88_RS22625 (G6M88_22635) | 26540..26839 | + | 300 | WP_065700840 | conjugal transfer protein TrbD | virB3 |
| G6M88_RS22630 (G6M88_22640) | 26850..29306 | + | 2457 | WP_065700841 | conjugal transfer protein TrbE | virb4 |
| G6M88_RS22635 (G6M88_22645) | 29278..30084 | + | 807 | WP_065700842 | P-type conjugative transfer protein TrbJ | virB5 |
| G6M88_RS22640 (G6M88_22650) | 30078..30254 | + | 177 | WP_065700843 | entry exclusion protein TrbK | - |
| G6M88_RS22645 (G6M88_22655) | 30298..31446 | + | 1149 | Protein_33 | P-type conjugative transfer protein TrbL | - |
| G6M88_RS22650 (G6M88_22660) | 31467..32129 | + | 663 | WP_065701003 | conjugal transfer protein TrbF | virB8 |
| G6M88_RS22655 (G6M88_22665) | 32147..32959 | + | 813 | WP_065700844 | P-type conjugative transfer protein TrbG | virB9 |
| G6M88_RS22660 (G6M88_22670) | 32962..33402 | + | 441 | WP_065700845 | conjugal transfer protein TrbH | - |
| G6M88_RS22665 (G6M88_22675) | 33415..34710 | + | 1296 | WP_065700846 | IncP-type conjugal transfer protein TrbI | virB10 |
| G6M88_RS22670 (G6M88_22680) | 34974..35678 | + | 705 | WP_065700847 | autoinducer binding domain-containing protein | - |
| G6M88_RS22675 (G6M88_22685) | 35681..36004 | - | 324 | WP_065700848 | transcriptional repressor TraM | - |
| G6M88_RS22680 (G6M88_22690) | 36213..36419 | + | 207 | WP_062653296 | helix-turn-helix domain-containing protein | - |
| G6M88_RS22685 (G6M88_22695) | 36499..38064 | - | 1566 | WP_065700849 | Fic family protein | - |
| G6M88_RS22690 (G6M88_22700) | 38288..38902 | - | 615 | WP_065700850 | TraH family protein | virB1 |
| G6M88_RS22695 (G6M88_22705) | 38919..40085 | - | 1167 | WP_065700851 | conjugal transfer protein TraB | - |
| G6M88_RS22700 (G6M88_22710) | 40075..40572 | - | 498 | WP_234791255 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 21423 | GenBank | NZ_CP049208 |
| Plasmid name | pW2_73_1 | Incompatibility group | - |
| Plasmid size | 289141 bp | Coordinate of oriT [Strand] | 44043..44076 [-] |
| Host baterium | Agrobacterium rubi strain W2/73 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |