Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120989 |
| Name | oriT_T808|p4 |
| Organism | Pararhizobium qamdonense strain T808 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP119569 (182286..182328 [-], 43 nt) |
| oriT length | 43 nt |
| IRs (inverted repeats) | 22..27, 31..36 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 43 nt
>oriT_T808|p4
GGATCCAAGGGCGCAATTATACGTCGCGATGCGACGCCTTGCG
GGATCCAAGGGCGCAATTATACGTCGCGATGCGACGCCTTGCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file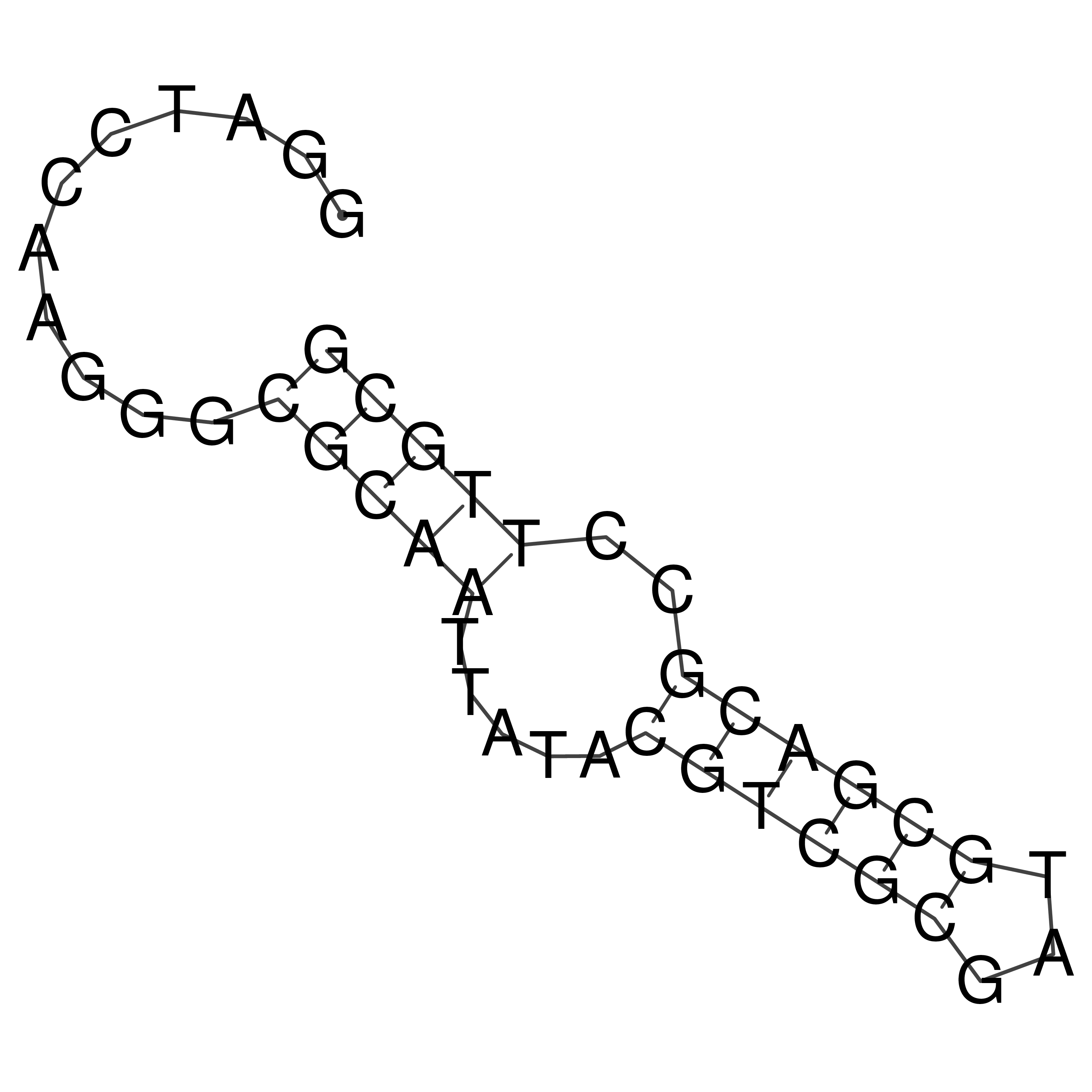
Relaxase
| ID | 13243 | GenBank | WP_276122413 |
| Name | traA_PYR65_RS29355_T808|p4 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123741.16 Da Isoelectric Point: 9.6720
>WP_276122413.1 Ti-type conjugative transfer relaxase TraA [Pararhizobium qamdonense]
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKHGLLHEEFMLPTDAPKWVRSLIADR
SVSGAVEAFWNRVEAFEKRSDAQLARDLTIALPLELSPEQNIALVRDFVEKHILAKGMIADWVYHDNPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGDDGQPVRTKSGKILYELWAGSTEDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYDKQGIDLEPTIHLGVGAKAIERKAEQQGVRPELERIELNEERRSENTRRILNNPAIVLDV
ITREKSVFDERDVAKVLHRYVDDPTVFQQLMLRIILNPEVLRLQRDTIEFATGEKVPARYSTQAMIRLEA
TMVRQALWLSNRDGHAVSEADLAATFRRHERLSQEQKTAIERIASPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAVVDRVGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANTRITGTRLKAEAVERLIGDWNHDYDQAKTTLILAHLRRDVRMLNVMAREKLVERGVVGEGHSFITADG
IRQFDAGDQIVFLKNETSLGVKNGMIGHVVEAAPNRIVAVTGEGEHRRHVVVEQRFYRNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFAFNGGLSKVLSRKNAKETTLDYERGQLY
REALRFAENRGLHIVQVARTLLRDRLDWTLRQKTKLVELGQRLAVFAERLGLHHPPKTQTMKEAAPMVAG
IKTFSGSVADTVEQKLDTDPALRQQWEEVSTRFAYVFADPETAFRAMNFDAVLVDKVTAKLALQTLTTAP
ASIGPLRGKTGILASKTEREDRRVADLNVPALKRDLEQYLRMRETAVQRLQADEQVLRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKTEIDVFNQAITSRFGERTLLTNAAREPSGKLFDKLSEG
KQPEQKERLTEAWPIMRASQQLAAHERTVQTLRQAEDLRLAPRQTPVMKQ
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKHGLLHEEFMLPTDAPKWVRSLIADR
SVSGAVEAFWNRVEAFEKRSDAQLARDLTIALPLELSPEQNIALVRDFVEKHILAKGMIADWVYHDNPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGDDGQPVRTKSGKILYELWAGSTEDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYDKQGIDLEPTIHLGVGAKAIERKAEQQGVRPELERIELNEERRSENTRRILNNPAIVLDV
ITREKSVFDERDVAKVLHRYVDDPTVFQQLMLRIILNPEVLRLQRDTIEFATGEKVPARYSTQAMIRLEA
TMVRQALWLSNRDGHAVSEADLAATFRRHERLSQEQKTAIERIASPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAVVDRVGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANTRITGTRLKAEAVERLIGDWNHDYDQAKTTLILAHLRRDVRMLNVMAREKLVERGVVGEGHSFITADG
IRQFDAGDQIVFLKNETSLGVKNGMIGHVVEAAPNRIVAVTGEGEHRRHVVVEQRFYRNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFAFNGGLSKVLSRKNAKETTLDYERGQLY
REALRFAENRGLHIVQVARTLLRDRLDWTLRQKTKLVELGQRLAVFAERLGLHHPPKTQTMKEAAPMVAG
IKTFSGSVADTVEQKLDTDPALRQQWEEVSTRFAYVFADPETAFRAMNFDAVLVDKVTAKLALQTLTTAP
ASIGPLRGKTGILASKTEREDRRVADLNVPALKRDLEQYLRMRETAVQRLQADEQVLRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKTEIDVFNQAITSRFGERTLLTNAAREPSGKLFDKLSEG
KQPEQKERLTEAWPIMRASQQLAAHERTVQTLRQAEDLRLAPRQTPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15521 | GenBank | WP_276122373 |
| Name | t4cp2_PYR65_RS29120_T808|p4 |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 91779.70 Da Isoelectric Point: 6.0937
>WP_276122373.1 conjugal transfer protein TrbE [Pararhizobium qamdonense]
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILARLG
SGWMIQVEAIRVPTGDYPTETESHFPDAVTRAIDDERRVHFQREKGHFESRHALILTWRPPEPRRSGLTR
YIYSEAASRSATYADRALESFLTSIREVEQYLANVVSIRRMMTRETPERGGYRIARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPMVESRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDSGRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPVAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGGSMLPLTLGLGGDHYQIGGDIGPSANGEVKALSFCPLAELSTDGDRAWASEWIEMLVALQGVIITP
DYRNAISRQLALMAGSRGRSLSDFVSGVQMRVIKDALHHYTVDGPMGQLLDAEQDGLALGEFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATASPKREYYVASPEGRRLFNM
ALGPVALSFVGATGKEDLKRIRSLHSEHGAAWPLHWLQQRGIAHAETFFPTS
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILARLG
SGWMIQVEAIRVPTGDYPTETESHFPDAVTRAIDDERRVHFQREKGHFESRHALILTWRPPEPRRSGLTR
YIYSEAASRSATYADRALESFLTSIREVEQYLANVVSIRRMMTRETPERGGYRIARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPMVESRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDSGRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPVAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGGSMLPLTLGLGGDHYQIGGDIGPSANGEVKALSFCPLAELSTDGDRAWASEWIEMLVALQGVIITP
DYRNAISRQLALMAGSRGRSLSDFVSGVQMRVIKDALHHYTVDGPMGQLLDAEQDGLALGEFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATASPKREYYVASPEGRRLFNM
ALGPVALSFVGATGKEDLKRIRSLHSEHGAAWPLHWLQQRGIAHAETFFPTS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15522 | GenBank | WP_276122416 |
| Name | traG_PYR65_RS29370_T808|p4 |
UniProt ID | _ |
| Length | 653 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 653 a.a. Molecular weight: 69901.69 Da Isoelectric Point: 9.4834
>WP_276122416.1 Ti-type conjugative transfer system protein TraG [Pararhizobium qamdonense]
MTANRLLLSIMPAVIIIAIMFATSGLEHRLAGFGSFAQTKLLLGRAGFALPYVAAAAIGVIALFAVNGSS
NIKAAALSVLAGNSAVIVIAVAREAIRLAGIASSLPAGQSVLAYTDPATMVGASASLFGGVFALRVAIKG
NAAFAQAGPTRIGGKRAVHGDANWMAMQEAGKLFPDAGGFVVGERYRVDRDSVAAVAFRTDNPQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGSGLVVLDPSSEVAPMVVEHRRKAGRKVIVLD
PGDPATGFNALDWIGRFGGTKEEDIVAVATWIMTDNARAASARDDFFRASAMQLLTALIADVCLSGHTDA
KDQTLRRVRSNLSEPEPKLRERLTRIYEQSESTFVRENVAVFVNMTPETFSGVYANAVKETHWLSYTNYA
ALVSGDSFSTDELATGVTDIFIALDLKVLESHPGLARVVIGSFLNAIYNRNGEVAGRTLFLLDEVARLGY
LRILETARDAGRKYGISLALIFQSIGQMREAYGGRDASSKWFESASWISFAAINDPETAEYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLTRRPLILPHEVLRMRGDEQIVFTAGNAPLRCGRAIWFRRKDMSASVG
ENRFHRQAIKGSEANETDPAPEN
MTANRLLLSIMPAVIIIAIMFATSGLEHRLAGFGSFAQTKLLLGRAGFALPYVAAAAIGVIALFAVNGSS
NIKAAALSVLAGNSAVIVIAVAREAIRLAGIASSLPAGQSVLAYTDPATMVGASASLFGGVFALRVAIKG
NAAFAQAGPTRIGGKRAVHGDANWMAMQEAGKLFPDAGGFVVGERYRVDRDSVAAVAFRTDNPQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGSGLVVLDPSSEVAPMVVEHRRKAGRKVIVLD
PGDPATGFNALDWIGRFGGTKEEDIVAVATWIMTDNARAASARDDFFRASAMQLLTALIADVCLSGHTDA
KDQTLRRVRSNLSEPEPKLRERLTRIYEQSESTFVRENVAVFVNMTPETFSGVYANAVKETHWLSYTNYA
ALVSGDSFSTDELATGVTDIFIALDLKVLESHPGLARVVIGSFLNAIYNRNGEVAGRTLFLLDEVARLGY
LRILETARDAGRKYGISLALIFQSIGQMREAYGGRDASSKWFESASWISFAAINDPETAEYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLTRRPLILPHEVLRMRGDEQIVFTAGNAPLRCGRAIWFRRKDMSASVG
ENRFHRQAIKGSEANETDPAPEN
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 128861..138554
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PYR65_RS29085 | 124245..125459 | - | 1215 | WP_276122366 | plasmid replication protein RepC | - |
| PYR65_RS29090 | 125614..126600 | - | 987 | WP_276122367 | plasmid partitioning protein RepB | - |
| PYR65_RS29095 | 126675..127862 | - | 1188 | WP_276122368 | plasmid partitioning protein RepA | - |
| PYR65_RS29100 | 128229..128864 | + | 636 | WP_276122369 | acyl-homoserine-lactone synthase | - |
| PYR65_RS29105 | 128861..129832 | + | 972 | WP_276122370 | P-type conjugative transfer ATPase TrbB | virB11 |
| PYR65_RS29110 | 129822..130226 | + | 405 | WP_276122371 | conjugal transfer pilin TrbC | virB2 |
| PYR65_RS29115 | 130219..130518 | + | 300 | WP_276122372 | conjugal transfer protein TrbD | virB3 |
| PYR65_RS29120 | 130522..132990 | + | 2469 | WP_276122373 | conjugal transfer protein TrbE | virb4 |
| PYR65_RS29125 | 133001..133771 | + | 771 | WP_276122550 | P-type conjugative transfer protein TrbJ | virB5 |
| PYR65_RS29130 | 133768..133995 | + | 228 | WP_276122374 | entry exclusion protein TrbK | - |
| PYR65_RS29135 | 133989..135179 | + | 1191 | WP_276122375 | P-type conjugative transfer protein TrbL | virB6 |
| PYR65_RS29140 | 135197..135874 | + | 678 | WP_276122376 | conjugal transfer protein TrbF | virB8 |
| PYR65_RS29145 | 135892..136743 | + | 852 | WP_276122551 | P-type conjugative transfer protein TrbG | virB9 |
| PYR65_RS29150 | 136743..137219 | + | 477 | WP_276122377 | conjugal transfer protein TrbH | - |
| PYR65_RS29155 | 137232..138554 | + | 1323 | WP_276122378 | IncP-type conjugal transfer protein TrbI | virB10 |
| PYR65_RS29160 | 138607..138861 | - | 255 | WP_276122379 | hypothetical protein | - |
| PYR65_RS29165 | 138895..140145 | - | 1251 | WP_276122380 | D-amino acid dehydrogenase | - |
| PYR65_RS29170 | 140171..141304 | - | 1134 | WP_276122552 | alanine racemase | - |
| PYR65_RS29175 | 141432..141896 | + | 465 | WP_276122381 | Lrp/AsnC family transcriptional regulator | - |
| PYR65_RS29180 | 142455..143348 | - | 894 | WP_276122382 | AraC family transcriptional regulator | - |
Host bacterium
| ID | 21418 | GenBank | NZ_CP119569 |
| Plasmid name | T808|p4 | Incompatibility group | - |
| Plasmid size | 248361 bp | Coordinate of oriT [Strand] | 182286..182328 [-] |
| Host baterium | Pararhizobium qamdonense strain T808 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |