Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120912 |
| Name | oriT_pHRC017 |
| Organism | Sinorhizobium meliloti |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_019313 (132455..132511 [-], 57 nt) |
| oriT length | 57 nt |
| IRs (inverted repeats) | 4..9, 23..28 (GGAAAA..TTTTCC) |
| Location of nic site | 36..37 |
| Conserved sequence flanking the nic site |
TCCTGCCTCT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 57 nt
>oriT_pHRC017
GCAGGAAAAGGGCGTAGCACATTTTTCCGTATCCTGCCTCTCCAAATTGTGAGGGGA
GCAGGAAAAGGGCGTAGCACATTTTTCCGTATCCTGCCTCTCCAAATTGTGAGGGGA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file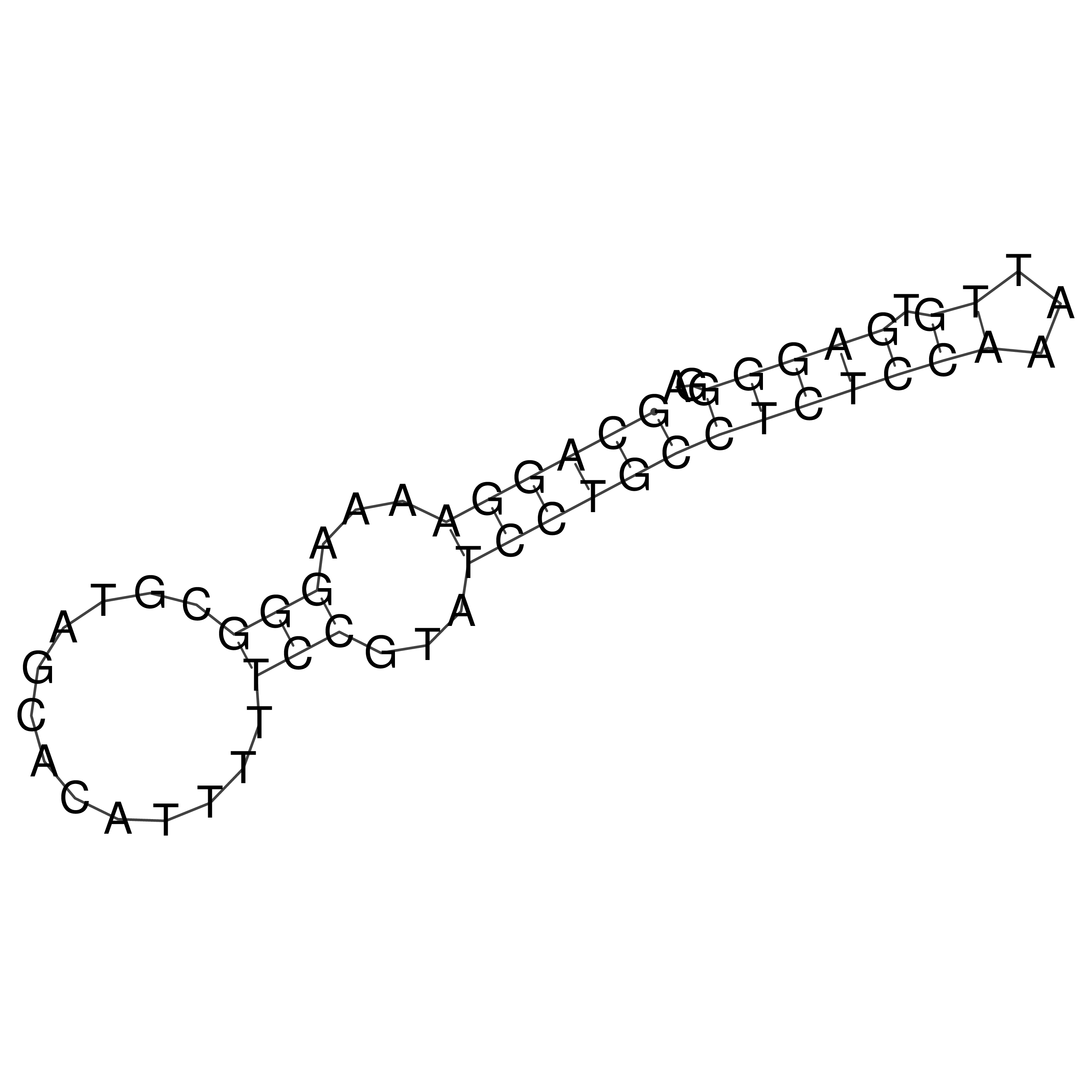
T4CP
| ID | 15471 | GenBank | WP_015061514 |
| Name | t4cp2_HTB76_RS01245_pHRC017 |
UniProt ID | _ |
| Length | 699 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 699 a.a. Molecular weight: 77736.65 Da Isoelectric Point: 9.8074
>WP_015061514.1 type IV secretory system conjugative DNA transfer family protein [Sinorhizobium meliloti]
MTKQLQAFYLLFCVGIAFVVWMLGYGLGLQLFYKDGRILETTITSNPFAPIQQFWHYKTSPALQKVALGS
MVPALLVAGLVAYIGLKPTSSPLGDAAFQDMASLRRAKWFRKQGHIFGRVGRNILRTKDDRHHLIIGPTR
SGKGAGYVIPNALMHEGSMIVTDLKGEVFKATAGYRRQNGSQVFLFAPGSEKTNSYNPLDFIRPERGNRT
TDIQNIASILVPENTESENSVWQATAQQVLAGVISYITESPFYKDRRNLAEVNSFFNSGVDLQALMKYIK
EKEPYLSKFTVESFNSYIALSERAAASALLDIQKAMRPFKNERIVAATNVTDMDLRAMKRRPISIYLAPN
ITDITLLRPLLTLFVQQVMDILTLEHDPNSLPVYFLLDEFRQLKRMDEIMTKLPYVAGYNIKLAFIIQDL
KNLDEIYGETSRHSLLGNCGYQLVLGANDQATAEYASRALGKRTIRYQSESRTIELMGLPRRTKVEQIRE
RDLMMPQEVRQMPETKMILLIEGQRPIFGEKLRFFQTQPFKSAEAFSQANIPQVPEVDYLAPKPVPATTP
EYAKGGDPSVEIPSLAPAKEEKPLTAAAAKAVPVKAERAADEKAAAPAKRTVNRQALRTNPKATAANTGG
AEASASLDATEARIKAIEEGLKPKAAQLKEVVETKAEKLGDKSPTKRRNILDIFSATVPDPVEVGVTAE
MTKQLQAFYLLFCVGIAFVVWMLGYGLGLQLFYKDGRILETTITSNPFAPIQQFWHYKTSPALQKVALGS
MVPALLVAGLVAYIGLKPTSSPLGDAAFQDMASLRRAKWFRKQGHIFGRVGRNILRTKDDRHHLIIGPTR
SGKGAGYVIPNALMHEGSMIVTDLKGEVFKATAGYRRQNGSQVFLFAPGSEKTNSYNPLDFIRPERGNRT
TDIQNIASILVPENTESENSVWQATAQQVLAGVISYITESPFYKDRRNLAEVNSFFNSGVDLQALMKYIK
EKEPYLSKFTVESFNSYIALSERAAASALLDIQKAMRPFKNERIVAATNVTDMDLRAMKRRPISIYLAPN
ITDITLLRPLLTLFVQQVMDILTLEHDPNSLPVYFLLDEFRQLKRMDEIMTKLPYVAGYNIKLAFIIQDL
KNLDEIYGETSRHSLLGNCGYQLVLGANDQATAEYASRALGKRTIRYQSESRTIELMGLPRRTKVEQIRE
RDLMMPQEVRQMPETKMILLIEGQRPIFGEKLRFFQTQPFKSAEAFSQANIPQVPEVDYLAPKPVPATTP
EYAKGGDPSVEIPSLAPAKEEKPLTAAAAKAVPVKAERAADEKAAAPAKRTVNRQALRTNPKATAANTGG
AEASASLDATEARIKAIEEGLKPKAAQLKEVVETKAEKLGDKSPTKRRNILDIFSATVPDPVEVGVTAE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 224364..234285
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTB76_RS01155 | 219548..219878 | + | 331 | Protein_235 | hypothetical protein | - |
| HTB76_RS01160 (pHRC017_0543) | 220378..220971 | - | 594 | WP_015061493 | hypothetical protein | - |
| HTB76_RS01165 (pHRC017_0545) | 220973..221377 | - | 405 | WP_153432028 | hypothetical protein | - |
| HTB76_RS01170 | 221378..222081 | - | 704 | Protein_238 | thermonuclease family protein | - |
| HTB76_RS01175 (pHRC017_0550) | 222078..222614 | - | 537 | WP_015061497 | thermonuclease family protein | - |
| HTB76_RS01180 (pHRC017_0551) | 222611..223219 | - | 609 | WP_015061498 | hypothetical protein | - |
| HTB76_RS01185 (pHRC017_0554) | 223442..224352 | + | 911 | Protein_241 | lytic transglycosylase domain-containing protein | - |
| HTB76_RS01190 (pHRC017_0557) | 224364..224702 | + | 339 | WP_015061502 | TrbC/VirB2 family protein | virB2 |
| HTB76_RS01195 (pHRC017_0558) | 224706..225005 | + | 300 | WP_014531077 | type IV secretion system protein VirB3 | virB3 |
| HTB76_RS01200 (pHRC017_0559) | 225008..227466 | + | 2459 | Protein_244 | VirB4 family type IV secretion/conjugal transfer ATPase | - |
| HTB76_RS01205 (pHRC017_0562) | 227463..228551 | + | 1089 | WP_153432035 | lytic transglycosylase domain-containing protein | - |
| HTB76_RS01210 (pHRC017_0564) | 228563..229264 | + | 702 | WP_015061507 | type IV secretion system protein | - |
| HTB76_RS01215 (pHRC017_0565) | 229254..229481 | + | 228 | WP_015061508 | hypothetical protein | - |
| HTB76_RS01220 (pHRC017_0567) | 229497..230525 | + | 1029 | WP_015061509 | type IV secretion system protein | virB6 |
| HTB76_RS01225 (pHRC017_0568) | 230564..231259 | + | 696 | WP_015061510 | virB8 family protein | virB8 |
| HTB76_RS01230 (pHRC017_0570) | 231256..232068 | + | 813 | WP_015061511 | P-type conjugative transfer protein VirB9 | virB9 |
| HTB76_RS01235 (pHRC017_0571) | 232065..233276 | + | 1212 | WP_015061512 | type IV secretion system protein VirB10 | virB10 |
| HTB76_RS01240 (pHRC017_0574) | 233257..234285 | + | 1029 | WP_015061513 | P-type DNA transfer ATPase VirB11 | virB11 |
| HTB76_RS01245 (pHRC017_0576) | 234282..236381 | + | 2100 | WP_015061514 | type IV secretory system conjugative DNA transfer family protein | - |
| HTB76_RS01250 (pHRC017_0578) | 236581..237489 | + | 909 | WP_015061515 | tyrosine-type recombinase/integrase | - |
| HTB76_RS01255 (pHRC017_0579) | 237493..238686 | + | 1194 | WP_015061516 | IS91 family transposase | - |
Host bacterium
| ID | 21341 | GenBank | NC_019313 |
| Plasmid name | pHRC017 | Incompatibility group | - |
| Plasmid size | 298356 bp | Coordinate of oriT [Strand] | 132455..132511 [-] |
| Host baterium | Sinorhizobium meliloti |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |