Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120836 |
| Name | oriT_p29A21006_A_KPC |
| Organism | Serratia liquefaciens strain 29A21CPO006 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_OQ821180 (43965..44559 [+], 595 nt) |
| oriT length | 595 nt |
| IRs (inverted repeats) | 569..574, 579..584 (ATGCCC..GGGCAT) 497..503, 508..514 (CCTCCCG..CGGGAGG) 433..438, 442..447 (GTTCGC..GCGAAC) 75..80, 86..91 (TGCTTT..AAAGCA) 69..74, 82..87 (TTACTT..AAGTAA) |
| Location of nic site | 294..295 |
| Conserved sequence flanking the nic site |
TCCTGCATCG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 595 nt
>oriT_p29A21006_A_KPC
TGTTAAATACATTGTAGTTTTGATAAAATCATTAGTATCTGGCATAAAGTTATCATTATCATCTTCTGTTACTTTGCTTTTAAGTAAAGCACTGGCAAGCAATTTCATGCCTTTAGCAACAGTTAAACCAGGCTCATCTGGATAAATAACCTTCAGACATTCAAGTAGATTATCATGCTGTTTTTCTTCTAGTCTGAAATTAATTTGCTTCATAAATCCTCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGAGTAAAATGCTAGCATTTTGCGGCGTTTGCCCTATCCTGCATCGCTTTGATCTAGCACTAGTCTTGAATTGTGCTTCAGCGTCGAGGAAAAAGCAAAACTGAAGGGCTGTCATGGCTGAAGGTTTCGCTTGTCTACGACCGGAAACATCTCAATGGCTGTGAGCTATGTGGGATAAGGTTCGCTACGCGAACGGTCTGGCAGGGGGCGCAAGCGCTGTCTTGCAGAGTATATAAAAAAGCACCTCCCGCAAACGGGAGGGCTTCGGCGATTCATGACGTGTGAAACTATTCAGGGTCTGGTGTGTCGCCTACCATGCCCTGACGGGCATCACACAACCAG
TGTTAAATACATTGTAGTTTTGATAAAATCATTAGTATCTGGCATAAAGTTATCATTATCATCTTCTGTTACTTTGCTTTTAAGTAAAGCACTGGCAAGCAATTTCATGCCTTTAGCAACAGTTAAACCAGGCTCATCTGGATAAATAACCTTCAGACATTCAAGTAGATTATCATGCTGTTTTTCTTCTAGTCTGAAATTAATTTGCTTCATAAATCCTCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGAGTAAAATGCTAGCATTTTGCGGCGTTTGCCCTATCCTGCATCGCTTTGATCTAGCACTAGTCTTGAATTGTGCTTCAGCGTCGAGGAAAAAGCAAAACTGAAGGGCTGTCATGGCTGAAGGTTTCGCTTGTCTACGACCGGAAACATCTCAATGGCTGTGAGCTATGTGGGATAAGGTTCGCTACGCGAACGGTCTGGCAGGGGGCGCAAGCGCTGTCTTGCAGAGTATATAAAAAAGCACCTCCCGCAAACGGGAGGGCTTCGGCGATTCATGACGTGTGAAACTATTCAGGGTCTGGTGTGTCGCCTACCATGCCCTGACGGGCATCACACAACCAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file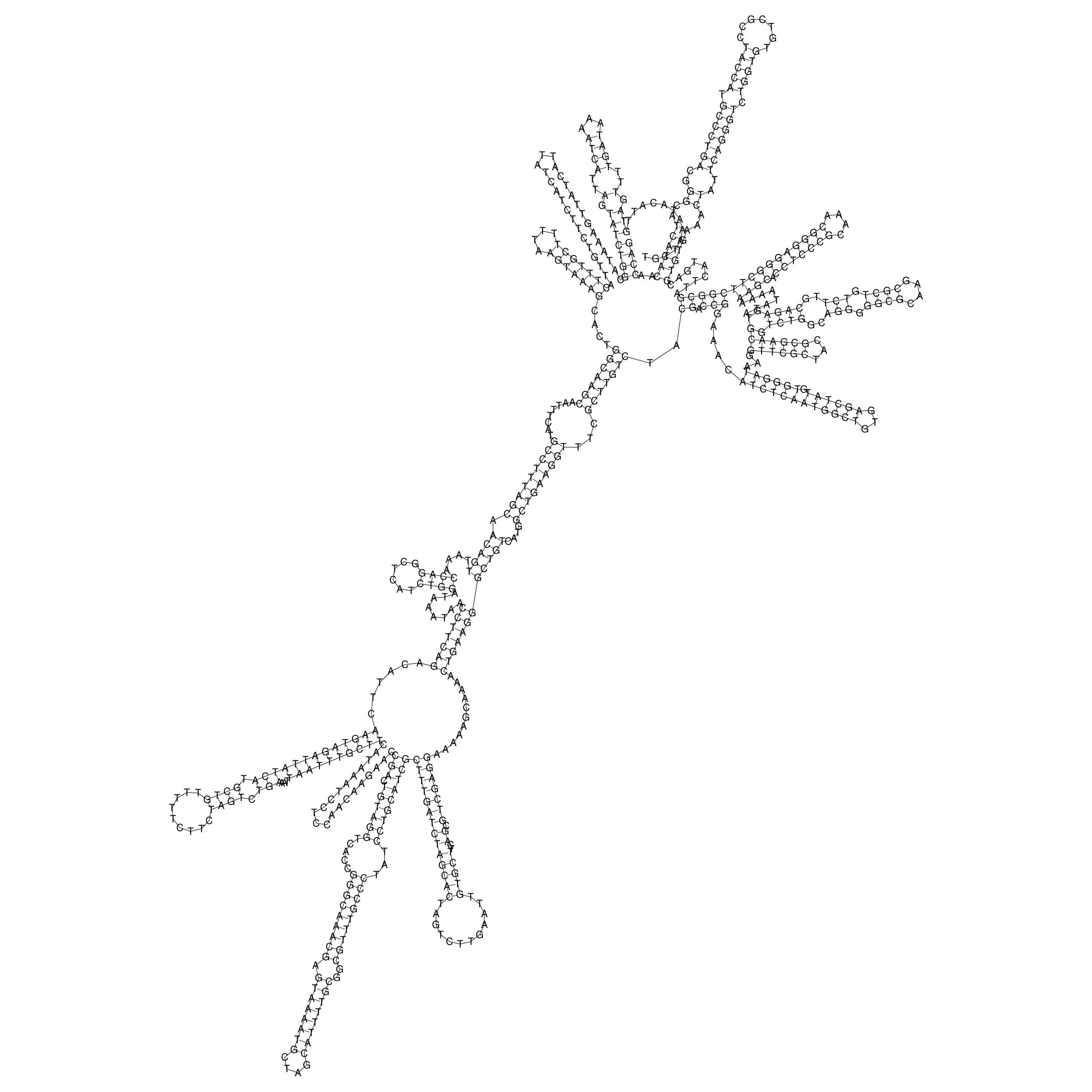
Relaxase
| ID | 13143 | GenBank | WP_044864205 |
| Name | mobP1_SLH58_RS00245_p29A21006_A_KPC |
UniProt ID | A0A221ZPJ2 |
| Length | 386 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 386 a.a. Molecular weight: 44352.85 Da Isoelectric Point: 10.6126
>WP_044864205.1 MULTISPECIES: MobP1 family relaxase [Enterobacterales]
MGVSVDKEYRVKRKSSEAGRKSAFAHKVKNGGKNYQRNVQERINRKGASKEVVVKISGGAITRQGIRNSI
DYMSRESELPVMSESGQVWKGDEIQEAKEHMVDRANDPQNVMNDKGKENKKVTQNIVFSPPVSAKVKPED
LLESVRKTMHKKYPNHRFVLGYHNDKKDHPHVHVVFRIRDEDGKRTDIRKKDLREIRTGFCEELKLKGYD
VKATHRQQQGLNQSIRDAHSTAPKRQKGVYEVVDVGYDHYQHDKTKPKQHFLKLKTLNKGVEKTYWGADF
GDLTSRENVKKGDLVKLKKLGQKEVKIPALDKHGVQHGWKTVHRNEWKLDNLGVKGIDRTPSSSKELVLN
SAAMLQKQQLQMKQFIQIKQAMLQNEQKMKIGIKLG
MGVSVDKEYRVKRKSSEAGRKSAFAHKVKNGGKNYQRNVQERINRKGASKEVVVKISGGAITRQGIRNSI
DYMSRESELPVMSESGQVWKGDEIQEAKEHMVDRANDPQNVMNDKGKENKKVTQNIVFSPPVSAKVKPED
LLESVRKTMHKKYPNHRFVLGYHNDKKDHPHVHVVFRIRDEDGKRTDIRKKDLREIRTGFCEELKLKGYD
VKATHRQQQGLNQSIRDAHSTAPKRQKGVYEVVDVGYDHYQHDKTKPKQHFLKLKTLNKGVEKTYWGADF
GDLTSRENVKKGDLVKLKKLGQKEVKIPALDKHGVQHGWKTVHRNEWKLDNLGVKGIDRTPSSSKELVLN
SAAMLQKQQLQMKQFIQIKQAMLQNEQKMKIGIKLG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A221ZPJ2 |
T4CP
| ID | 15424 | GenBank | WP_044715174 |
| Name | t4cp2_SLH58_RS00185_p29A21006_A_KPC |
UniProt ID | _ |
| Length | 611 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 611 a.a. Molecular weight: 69245.98 Da Isoelectric Point: 7.1493
>WP_044715174.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacterales]
MSLKLPDKGQWLFIGFVMCLFTYYAGSVAVYFFNGKTPLFIWKNFDSMLLWRLMTESNIRSDIRITAMPA
LLSGLAASVIVPVFIIWQLNKKDFSLYGDAKFASDDDLKKSKLLKWEKENDDDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREVILQNKVFLLDPFNSKTHKFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYAPDWLNEFNLKPIFSI
GSVVDLYSNIDRENVLANREDFEGIAGLDETALFHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDISIYVGVNAEDMSLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAKKIS
EKLGYTTAKSTGSSKTSGRSTSKSNSESDAQRALVLPQELGTLEFSEEFIILKGENPVKAKKALYYLDSY
FMDRLMLVSPKLAALTSSLNKTKNIFGVKGLKYPSKEKMLSVGELESEVLL
MSLKLPDKGQWLFIGFVMCLFTYYAGSVAVYFFNGKTPLFIWKNFDSMLLWRLMTESNIRSDIRITAMPA
LLSGLAASVIVPVFIIWQLNKKDFSLYGDAKFASDDDLKKSKLLKWEKENDDDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREVILQNKVFLLDPFNSKTHKFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYAPDWLNEFNLKPIFSI
GSVVDLYSNIDRENVLANREDFEGIAGLDETALFHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDISIYVGVNAEDMSLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAKKIS
EKLGYTTAKSTGSSKTSGRSTSKSNSESDAQRALVLPQELGTLEFSEEFIILKGENPVKAKKALYYLDSY
FMDRLMLVSPKLAALTSSLNKTKNIFGVKGLKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 31440..41395
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SLH58_RS00155 (AMFCNDPM_00028) | 26999..27772 | - | 774 | WP_044715185 | hypothetical protein | - |
| SLH58_RS00160 (AMFCNDPM_00029) | 27787..28257 | - | 471 | WP_044715183 | thermonuclease family protein | - |
| SLH58_RS00165 (AMFCNDPM_00030) | 28254..28574 | - | 321 | WP_044715215 | hypothetical protein | - |
| SLH58_RS00170 (AMFCNDPM_00031) | 28595..28798 | - | 204 | WP_044715181 | hypothetical protein | - |
| SLH58_RS00175 (AMFCNDPM_00032) | 28867..29205 | - | 339 | WP_225622178 | hypothetical protein | - |
| SLH58_RS00180 (AMFCNDPM_00033) | 29210..29605 | - | 396 | WP_044715177 | cag pathogenicity island Cag12 family protein | - |
| SLH58_RS00185 (AMFCNDPM_00034) | 29602..31437 | - | 1836 | WP_044715174 | type IV secretory system conjugative DNA transfer family protein | - |
| SLH58_RS00190 (AMFCNDPM_00035) | 31440..32471 | - | 1032 | WP_044715211 | P-type DNA transfer ATPase VirB11 | virB11 |
| SLH58_RS00195 (AMFCNDPM_00036) | 32495..33706 | - | 1212 | WP_044715172 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| SLH58_RS00200 (AMFCNDPM_00037) | 33703..34632 | - | 930 | WP_044715170 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| SLH58_RS00205 (AMFCNDPM_00038) | 34637..35365 | - | 729 | WP_044715168 | virB8 family protein | virB8 |
| SLH58_RS00210 (AMFCNDPM_00039) | 35605..36660 | - | 1056 | WP_117242821 | type IV secretion system protein | virB6 |
| SLH58_RS00215 (AMFCNDPM_00040) | 36672..36929 | - | 258 | WP_044715163 | EexN family lipoprotein | - |
| SLH58_RS00220 (AMFCNDPM_00041) | 36939..37685 | - | 747 | WP_044715162 | type IV secretion system protein | - |
| SLH58_RS00225 (AMFCNDPM_00042) | 37696..40455 | - | 2760 | WP_044715159 | VirB3 family type IV secretion system protein | virb4 |
| SLH58_RS00230 (AMFCNDPM_00043) | 40477..40767 | - | 291 | WP_044715156 | TrbC/VirB2 family protein | virB2 |
| SLH58_RS00235 (AMFCNDPM_00044) | 40751..41395 | - | 645 | WP_044715155 | lytic transglycosylase domain-containing protein | virB1 |
| SLH58_RS00240 (AMFCNDPM_00046) | 41619..42110 | - | 492 | WP_044715152 | transcription termination/antitermination NusG family protein | - |
| SLH58_RS00245 (AMFCNDPM_00047) | 42466..43626 | - | 1161 | WP_044864205 | MobP1 family relaxase | - |
| SLH58_RS00250 (AMFCNDPM_00048) | 43629..44177 | - | 549 | WP_044715148 | DNA distortion polypeptide 1 | - |
| SLH58_RS00255 (AMFCNDPM_00049) | 44505..44762 | - | 258 | WP_044715147 | hypothetical protein | - |
| SLH58_RS00260 (AMFCNDPM_00050) | 44778..44927 | - | 150 | WP_162852120 | hypothetical protein | - |
| SLH58_RS00265 (AMFCNDPM_00051) | 45005..45346 | - | 342 | WP_059347451 | hypothetical protein | - |
| SLH58_RS00270 (AMFCNDPM_00052) | 45443..45682 | - | 240 | WP_044715143 | hypothetical protein | - |
| SLH58_RS00275 (AMFCNDPM_00053) | 45675..45956 | - | 282 | WP_048235114 | hypothetical protein | - |
Host bacterium
| ID | 21265 | GenBank | NZ_OQ821180 |
| Plasmid name | p29A21006_A_KPC | Incompatibility group | IncX5 |
| Plasmid size | 56021 bp | Coordinate of oriT [Strand] | 43965..44559 [+] |
| Host baterium | Serratia liquefaciens strain 29A21CPO006 |
Cargo genes
| Drug resistance gene | blaKPC-2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |