Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120835 |
| Name | oriT_p31A17006_B_KPC |
| Organism | Serratia marcescens strain 31A17CRGN006 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_OQ821183 (57552..57652 [-], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 80..85, 91..96 (AAAAAA..TTTTTT) 20..26, 38..44 (TAAATCA..TGATTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_p31A17006_B_KPC
TATTTATTTTTTTATCTTTTAAATCAGTATGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
TATTTATTTTTTTATCTTTTAAATCAGTATGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file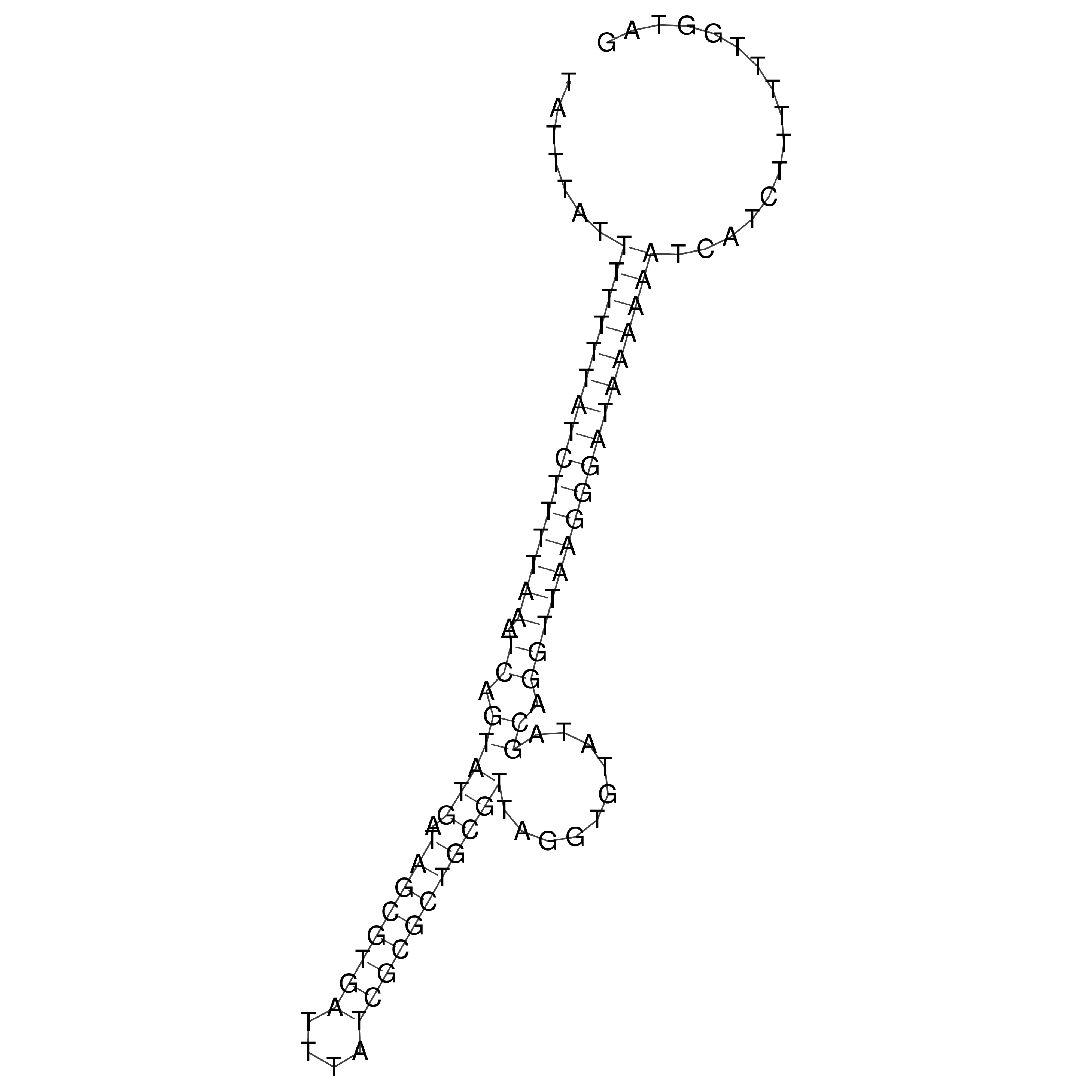
T4CP
| ID | 15423 | GenBank | WP_319382229 |
| Name | t4cp2_SLH58_RS00360_p31A17006_B_KPC |
UniProt ID | _ |
| Length | 506 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 506 a.a. Molecular weight: 57296.36 Da Isoelectric Point: 9.5206
>WP_319382229.1 type IV secretion system DNA-binding domain-containing protein [Serratia marcescens]
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVMLVFRDLTSAAQM
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVMLVFRDLTSAAQM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 12514..22839
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SLH58_RS00045 (CBNHEBFE_00008) | 8706..9815 | + | 1110 | WP_000842134 | S-(hydroxymethyl)glutathione dehydrogenase/class III alcohol dehydrogenase | - |
| SLH58_RS00050 (CBNHEBFE_00009) | 9837..10259 | + | 423 | Protein_9 | alpha/beta hydrolase-fold protein | - |
| SLH58_RS00055 (CBNHEBFE_00010) | 10305..11009 | - | 705 | WP_001067855 | IS6-like element IS26 family transposase | - |
| SLH58_RS00060 (CBNHEBFE_00011) | 11074..11253 | - | 180 | Protein_11 | helix-turn-helix domain-containing protein | - |
| SLH58_RS00065 | 11520..11807 | + | 288 | Protein_12 | DUF6710 family protein | - |
| SLH58_RS00070 (CBNHEBFE_00012) | 11981..12514 | - | 534 | WP_000792636 | phospholipase D family protein | - |
| SLH58_RS00075 (CBNHEBFE_00013) | 12514..13509 | - | 996 | WP_000128596 | ATPase, T2SS/T4P/T4SS family | virB11 |
| SLH58_RS00080 (CBNHEBFE_00014) | 13551..14711 | - | 1161 | WP_000101710 | type IV secretion system protein VirB10 | virB10 |
| SLH58_RS00085 (CBNHEBFE_00015) | 14711..15595 | - | 885 | WP_000735066 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| SLH58_RS00090 (CBNHEBFE_00016) | 15606..16304 | - | 699 | WP_000646594 | virB8 family protein | virB8 |
| SLH58_RS00095 | 16294..16452 | - | 159 | WP_012561180 | hypothetical protein | - |
| SLH58_RS00100 (CBNHEBFE_00017) | 16523..17563 | - | 1041 | WP_001749958 | type IV secretion system protein | virB6 |
| SLH58_RS00105 (CBNHEBFE_00018) | 17579..17806 | - | 228 | WP_001749959 | IncN-type entry exclusion lipoprotein EexN | - |
| SLH58_RS00110 (CBNHEBFE_00019) | 17814..18578 | - | 765 | WP_013149461 | type IV secretion system protein | virB5 |
| SLH58_RS00115 (CBNHEBFE_00020) | 18545..21145 | - | 2601 | WP_001749961 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| SLH58_RS00120 (CBNHEBFE_00021) | 21145..21462 | - | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| SLH58_RS00125 (CBNHEBFE_00022) | 21512..21805 | - | 294 | WP_001749962 | hypothetical protein | virB2 |
| SLH58_RS00130 (CBNHEBFE_00023) | 21815..22096 | - | 282 | WP_000440698 | transcriptional repressor KorA | - |
| SLH58_RS00135 (CBNHEBFE_00024) | 22105..22839 | - | 735 | WP_001749963 | lytic transglycosylase domain-containing protein | virB1 |
| SLH58_RS00140 (CBNHEBFE_00025) | 22882..23253 | + | 372 | WP_011867773 | H-NS family nucleoid-associated regulatory protein | - |
| SLH58_RS00145 (CBNHEBFE_00026) | 23269..23613 | + | 345 | WP_001749964 | hypothetical protein | - |
| SLH58_RS00150 (CBNHEBFE_00027) | 23610..23924 | + | 315 | WP_001749965 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| SLH58_RS00155 (CBNHEBFE_00028) | 24038..24271 | + | 234 | WP_001191790 | hypothetical protein | - |
| SLH58_RS00160 (CBNHEBFE_00029) | 24321..24968 | + | 648 | WP_012561184 | restriction endonuclease | - |
| SLH58_RS00165 (CBNHEBFE_00030) | 24973..25179 | + | 207 | WP_001749967 | hypothetical protein | - |
| SLH58_RS00170 (CBNHEBFE_00031) | 25190..25462 | - | 273 | Protein_33 | IS1 family transposase | - |
| SLH58_RS00175 (CBNHEBFE_00032) | 25561..26994 | + | 1434 | WP_001288432 | DNA cytosine methyltransferase | - |
Host bacterium
| ID | 21264 | GenBank | NZ_OQ821183 |
| Plasmid name | p31A17006_B_KPC | Incompatibility group | IncN |
| Plasmid size | 88281 bp | Coordinate of oriT [Strand] | 57552..57652 [-] |
| Host baterium | Serratia marcescens strain 31A17CRGN006 |
Cargo genes
| Drug resistance gene | blaKPC-3, tet(D), dfrA14, sul2, aph(3'')-Ib, aph(6)-Id, blaOXA-9, ant(3'')-Ia, aac(6')-Ib |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |