Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120829 |
| Name | oriT_pRlX4 |
| Organism | Rhizobium beringeri strain TP13 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP140877 (995218..995246 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRlX4
AGGGCGCAATATACGTCGCTATCGCGACG
AGGGCGCAATATACGTCGCTATCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file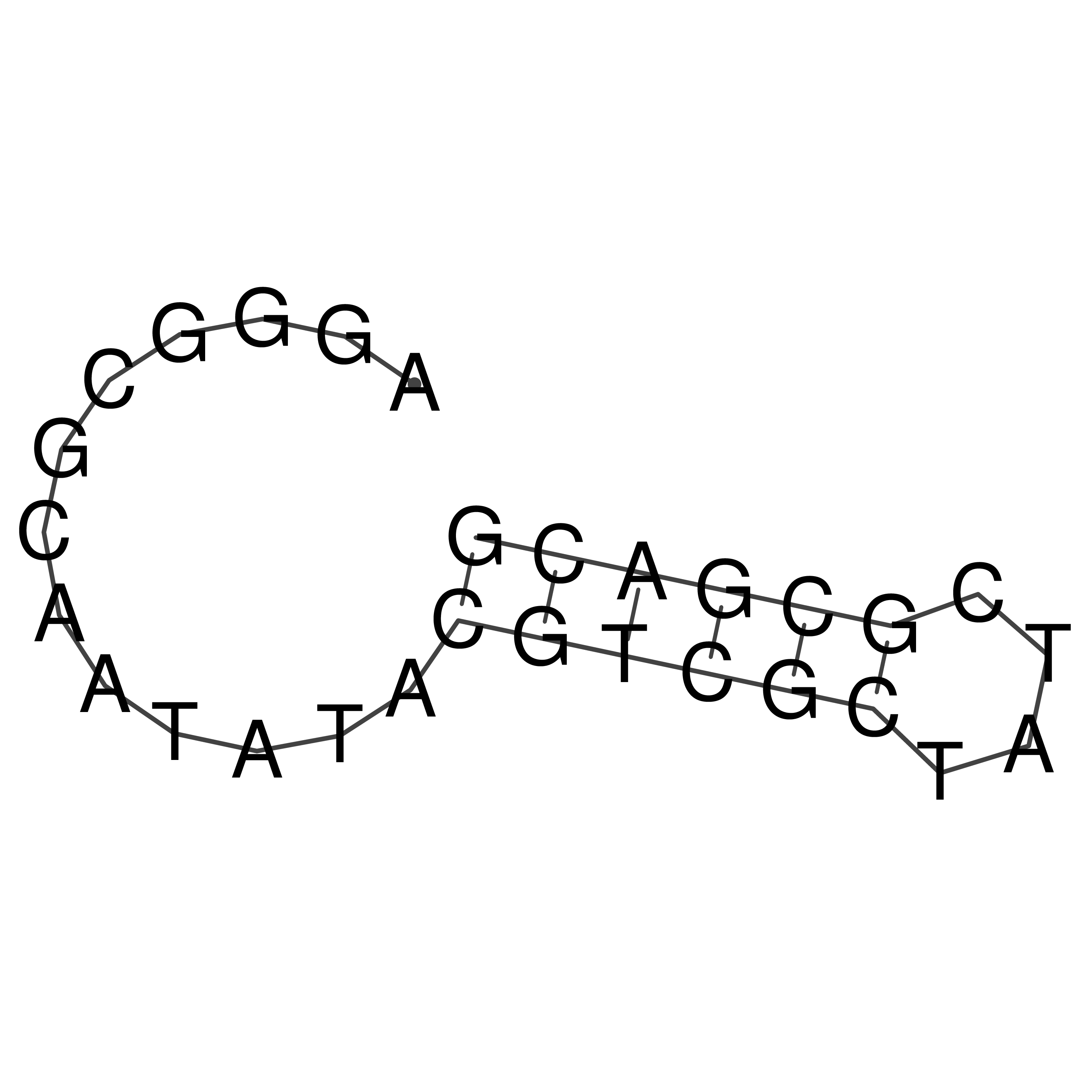
Relaxase
| ID | 13139 | GenBank | WP_327212794 |
| Name | traA_U8Q06_RS36050_pRlX4 |
UniProt ID | _ |
| Length | 1535 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1535 a.a. Molecular weight: 169991.82 Da Isoelectric Point: 7.1785
>WP_327212794.1 Ti-type conjugative transfer relaxase TraA [Rhizobium beringeri]
MAIMFVRAQVISRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGASELVHEELALPDQMPTWLRTAIDG
RSVAGASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDRDG
NPHIHLMTTLRPLTEEGFGQKKVAVTGDDGAPLRVVTPDRPKGRIVYRLWAGDKETMKAWKIAWADTANR
HLALAGHEVRLDGRSYAEQGLDGIAQTHLGPEKAALVRKGRELHFAPADLARRQEMADRLLAEPELLLKQ
LSNERSTFDERDIAKALHRYVDDPAVFANIRARLMVSDDLIMLKPQQLDAETGKASEPAIFTTRQILRIE
YDMARSAQVLAEYRGFAVSDRVVAAAISNVETRDPQKSFHLDPEQVDAVRHVTRDNGIAAVVGLAGAGKS
TLLAAARVAWESDGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELTWASGRELLERGNVLVIDEAGMVS
SQQMARVLDIAEQAEAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQQWAREASRLFARGE
VEQGLDAYARHGHLVEAGTRDEVIARIVGDWSEARKLAIETSVRQGNDGRLRGDELLVLAHTNEDVKRLN
EALRSVMSQEGALSENRTFRTERGARDFAAGDRIIFLENARFIEPRAPRLGPQHVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEAKPEWGRKPHVDHAAGVTGALVETGQAKFRPSDEDADDSPYADVRTDDGTIHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVTIKVPVVDEQTGQKRFEEREVDRNVWTAKRIETSEERQQRIEQESHRP
DLFKQLVERLSRSGAKTTTLDFESEASYRAQAQDFARRRGLDHLSLAAAAVEENLSRRWAGIAERRKQLA
KLWERAGVAFGFTIERERRVAYNEERAEPQPVEADVVGSRYLIPPATGFARSADEDALLAQLASPAWKER
EAILRPLLAKIYRDPDAALVALNALSSDTSIEPRRLADDLANAPDRLGRLRGSDLIIDGRAARDERNTAI
TALRDLLPLARAHATEFRRNGERFDLGEQRRRTHMSLPIPALSEKAMARLMEIEAVRRQGGDDAYKTAFA
LATEDRSIVQEVKAVSEALSARFGWSAFTAKADAVAERNMIERMPEDLTSISGEKLTRLFEAVKRFADEQ
HLVERRDRSKIVAAASADPQKENVPMLAAVTEFKTAVDEEARSRALATPLYRHQRAALAAVASTIWRDPA
GAVGKIEELLQKGFAGERIAAAVTNDPAAYGALRGSDRLVDRMLAAGRERKEVVQAVPEAGARLRALGSA
YVNLLDAERQATTEERRRMAVAIPGLSTAAEDVLMRLTAEAKNNGRKLSGSAASLGPDIRREFAAVSSAL
DERFGRSAIIRGEKDLSNRVPPAQHRAFEVMQEKLKVLQQTVRRESSEQIISERRQRTTNRSIDL
MAIMFVRAQVISRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGASELVHEELALPDQMPTWLRTAIDG
RSVAGASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDRDG
NPHIHLMTTLRPLTEEGFGQKKVAVTGDDGAPLRVVTPDRPKGRIVYRLWAGDKETMKAWKIAWADTANR
HLALAGHEVRLDGRSYAEQGLDGIAQTHLGPEKAALVRKGRELHFAPADLARRQEMADRLLAEPELLLKQ
LSNERSTFDERDIAKALHRYVDDPAVFANIRARLMVSDDLIMLKPQQLDAETGKASEPAIFTTRQILRIE
YDMARSAQVLAEYRGFAVSDRVVAAAISNVETRDPQKSFHLDPEQVDAVRHVTRDNGIAAVVGLAGAGKS
TLLAAARVAWESDGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELTWASGRELLERGNVLVIDEAGMVS
SQQMARVLDIAEQAEAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQQWAREASRLFARGE
VEQGLDAYARHGHLVEAGTRDEVIARIVGDWSEARKLAIETSVRQGNDGRLRGDELLVLAHTNEDVKRLN
EALRSVMSQEGALSENRTFRTERGARDFAAGDRIIFLENARFIEPRAPRLGPQHVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEAKPEWGRKPHVDHAAGVTGALVETGQAKFRPSDEDADDSPYADVRTDDGTIHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVTIKVPVVDEQTGQKRFEEREVDRNVWTAKRIETSEERQQRIEQESHRP
DLFKQLVERLSRSGAKTTTLDFESEASYRAQAQDFARRRGLDHLSLAAAAVEENLSRRWAGIAERRKQLA
KLWERAGVAFGFTIERERRVAYNEERAEPQPVEADVVGSRYLIPPATGFARSADEDALLAQLASPAWKER
EAILRPLLAKIYRDPDAALVALNALSSDTSIEPRRLADDLANAPDRLGRLRGSDLIIDGRAARDERNTAI
TALRDLLPLARAHATEFRRNGERFDLGEQRRRTHMSLPIPALSEKAMARLMEIEAVRRQGGDDAYKTAFA
LATEDRSIVQEVKAVSEALSARFGWSAFTAKADAVAERNMIERMPEDLTSISGEKLTRLFEAVKRFADEQ
HLVERRDRSKIVAAASADPQKENVPMLAAVTEFKTAVDEEARSRALATPLYRHQRAALAAVASTIWRDPA
GAVGKIEELLQKGFAGERIAAAVTNDPAAYGALRGSDRLVDRMLAAGRERKEVVQAVPEAGARLRALGSA
YVNLLDAERQATTEERRRMAVAIPGLSTAAEDVLMRLTAEAKNNGRKLSGSAASLGPDIRREFAAVSSAL
DERFGRSAIIRGEKDLSNRVPPAQHRAFEVMQEKLKVLQQTVRRESSEQIISERRQRTTNRSIDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15419 | GenBank | WP_327212793 |
| Name | traG_U8Q06_RS36035_pRlX4 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 70453.33 Da Isoelectric Point: 9.4055
>WP_327212793.1 Ti-type conjugative transfer system protein TraG [Rhizobium beringeri]
MALKGRLHPSLLLVFVPIAVTAITVYITGWRWTALAHGLSGKTEYWFLRAAPVPALLSGPLAGLLTVWAL
PLHRRRPIAIASFLCFLVVTGFYALREYGRLVPSVERGVVTWNQALSYLDFIAVIGALAGFMAVAVAARI
SVVVPEAVKRARGGTFGDADWLSMSAAAKLFPPEGEIVVGERYRVDKEIVHQLPFDPKDRSTWGQGGQAP
LLTYNQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVVEHRTRILGREVMVLDPA
NPIMGFNVLDGIEHSKRKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPECSGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSETAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSSDIVTGKTDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGGFSRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKGGGTRKSERVNFQRRPLIMPHEITQSMRKDEQIIIVPGHSPIRCGRAIYFRRKDMDAQAK
ANRFVRR
MALKGRLHPSLLLVFVPIAVTAITVYITGWRWTALAHGLSGKTEYWFLRAAPVPALLSGPLAGLLTVWAL
PLHRRRPIAIASFLCFLVVTGFYALREYGRLVPSVERGVVTWNQALSYLDFIAVIGALAGFMAVAVAARI
SVVVPEAVKRARGGTFGDADWLSMSAAAKLFPPEGEIVVGERYRVDKEIVHQLPFDPKDRSTWGQGGQAP
LLTYNQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVVEHRTRILGREVMVLDPA
NPIMGFNVLDGIEHSKRKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPECSGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSETAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSSDIVTGKTDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGGFSRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKGGGTRKSERVNFQRRPLIMPHEITQSMRKDEQIIIVPGHSPIRCGRAIYFRRKDMDAQAK
ANRFVRR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 691557..701155
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| U8Q06_RS34580 (U8Q06_34580) | 687640..688638 | + | 999 | WP_327212694 | ABC transporter substrate-binding protein | - |
| U8Q06_RS34585 (U8Q06_34585) | 688622..690397 | + | 1776 | WP_327212695 | adenylate/guanylate cyclase domain-containing protein | - |
| U8Q06_RS34590 (U8Q06_34590) | 690573..690926 | - | 354 | WP_128404288 | MarR family transcriptional regulator | - |
| U8Q06_RS34595 (U8Q06_34595) | 691018..691557 | + | 540 | WP_327212696 | hypothetical protein | - |
| U8Q06_RS34600 (U8Q06_34600) | 691557..692219 | + | 663 | WP_327212697 | lytic transglycosylase domain-containing protein | virB1 |
| U8Q06_RS34605 (U8Q06_34605) | 692216..692515 | + | 300 | WP_128404291 | TrbC/VirB2 family protein | virB2 |
| U8Q06_RS34610 (U8Q06_34610) | 692521..692859 | + | 339 | WP_327212698 | type IV secretion system protein VirB3 | virB3 |
| U8Q06_RS34615 (U8Q06_34615) | 692852..695218 | + | 2367 | WP_327212699 | VirB4 family type IV secretion system protein | virb4 |
| U8Q06_RS34620 (U8Q06_34620) | 695254..695916 | + | 663 | WP_327212855 | P-type DNA transfer protein VirB5 | virB5 |
| U8Q06_RS34625 (U8Q06_34625) | 695913..696137 | + | 225 | WP_128410222 | EexN family lipoprotein | - |
| U8Q06_RS34630 (U8Q06_34630) | 696141..697061 | + | 921 | WP_327212700 | type IV secretion system protein | virB6 |
| U8Q06_RS34635 (U8Q06_34635) | 697114..697398 | + | 285 | WP_233943224 | hypothetical protein | - |
| U8Q06_RS34640 (U8Q06_34640) | 697400..698071 | + | 672 | WP_327212701 | virB8 family protein | virB8 |
| U8Q06_RS34645 (U8Q06_34645) | 698068..698925 | + | 858 | WP_327212702 | P-type conjugative transfer protein VirB9 | virB9 |
| U8Q06_RS34650 (U8Q06_34650) | 698935..700107 | + | 1173 | WP_327212703 | type IV secretion system protein VirB10 | virB10 |
| U8Q06_RS34655 (U8Q06_34655) | 700115..701155 | + | 1041 | WP_327212704 | P-type DNA transfer ATPase VirB11 | virB11 |
| U8Q06_RS34660 (U8Q06_34660) | 701249..701704 | + | 456 | WP_222313228 | NUDIX hydrolase | - |
| U8Q06_RS34665 (U8Q06_34665) | 701701..702723 | + | 1023 | WP_327212705 | PHB depolymerase family esterase | - |
| U8Q06_RS34670 (U8Q06_34670) | 702806..702958 | + | 153 | Protein_648 | P-type DNA transfer ATPase VirB11 | - |
| U8Q06_RS34675 (U8Q06_34675) | 703177..703392 | - | 216 | WP_130832663 | DUF3072 domain-containing protein | - |
| U8Q06_RS34680 (U8Q06_34680) | 703639..704550 | - | 912 | WP_130660240 | LysR family transcriptional regulator | - |
| U8Q06_RS34685 (U8Q06_34685) | 704650..705411 | + | 762 | WP_327212706 | SDR family NAD(P)-dependent oxidoreductase | - |
Host bacterium
| ID | 21258 | GenBank | NZ_CP140877 |
| Plasmid name | pRlX4 | Incompatibility group | - |
| Plasmid size | 1103503 bp | Coordinate of oriT [Strand] | 995218..995246 [+] |
| Host baterium | Rhizobium beringeri strain TP13 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | - |