Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120824 |
| Name | oriT_pRlX1 |
| Organism | Rhizobium beringeri strain TP24 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP140852 (183925..183962 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pRlX1
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file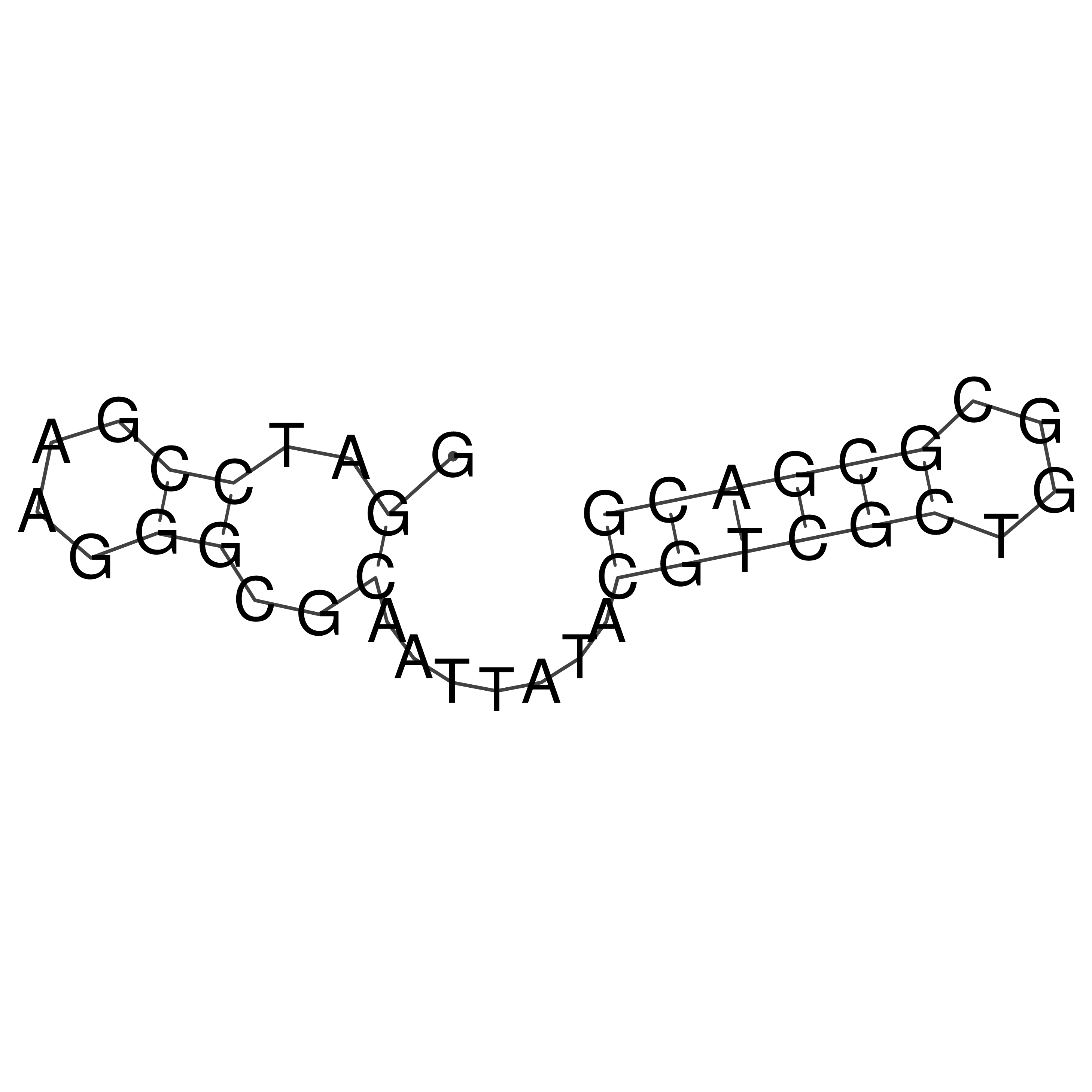
Relaxase
| ID | 13134 | GenBank | WP_128401822 |
| Name | traA_U8P69_RS24910_pRlX1 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123009.21 Da Isoelectric Point: 9.7914
>WP_128401822.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVVPADAPEWLRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTVEQNIALMRDFVERHITAQGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDDKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRVDGRSFEKQGIELEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVPEAVLQATFARHSRLSDEQRTAIEHIAGTTRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDRCVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMLGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLAYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYE
KSALYRQALRFAEARGLHLMNVARTIAHDRLQWAVRQSSRLADLGARLAAIGKALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPEAAFKAINVDAMLKDQSVAQS
TLARIAGKPESFGALKGNTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPSAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDAGG
KTFETVTTGMTPGQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVVPADAPEWLRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTVEQNIALMRDFVERHITAQGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDDKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRVDGRSFEKQGIELEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVPEAVLQATFARHSRLSDEQRTAIEHIAGTTRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDRCVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMLGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLAYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYE
KSALYRQALRFAEARGLHLMNVARTIAHDRLQWAVRQSSRLADLGARLAAIGKALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPEAAFKAINVDAMLKDQSVAQS
TLARIAGKPESFGALKGNTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPSAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDAGG
KTFETVTTGMTPGQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15413 | GenBank | WP_128411486 |
| Name | t4cp2_U8P69_RS24770_pRlX1 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 91617.41 Da Isoelectric Point: 6.1236
>WP_128411486.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVSLRLFRHTAPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESSTDFERNEMSRQINAIMSRLG
SGWMIQVEAVRLATVDYATAGKSHFPDRVTRAIDEERRAHFEREQGHFESRHAIIATYRPTEQRRSRLSK
YLYSDEGSRRRSYADTVLFLFRNAIRELEQYLANIVSIRRMLTREIAERGGARVARYDELLQFIRFCVTG
ENHPVRLPDIAMYLDWVATAELQHGLSPRVESRFLGVVAIDGLPSESWPGILNGLDAMPLSYRWSSRFIF
LDAEEAKVRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEEAIAEASSQLVAYGYYTPVVVL
FENDPELLNEKVEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTRNLSDLIPLNTVWS
GNPQAPCPFYPPGSPPLMQVATGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGRSMLPLTLAAGGDYYDVGGDEDTSLSFCPLAELSSDADRAWASEWIESLVAMQGITITPDHRNAIS
RQIGLMAVAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAETDGLALGLFQAFEIEQVMSMGERN
LVPVLTYLFRRIEKRLTGAPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAERSGI
IDVLKESCPTKICLPNGAAREPGTREFYERIGFNQRQIQIVASAIPKREYYVASPDGRRLFDMALGPLTL
SFVGASSKDDLKRIRALHEEHGPEWPLHWLQQRGIKDAASLLNDT
MVSLRLFRHTAPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESSTDFERNEMSRQINAIMSRLG
SGWMIQVEAVRLATVDYATAGKSHFPDRVTRAIDEERRAHFEREQGHFESRHAIIATYRPTEQRRSRLSK
YLYSDEGSRRRSYADTVLFLFRNAIRELEQYLANIVSIRRMLTREIAERGGARVARYDELLQFIRFCVTG
ENHPVRLPDIAMYLDWVATAELQHGLSPRVESRFLGVVAIDGLPSESWPGILNGLDAMPLSYRWSSRFIF
LDAEEAKVRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEEAIAEASSQLVAYGYYTPVVVL
FENDPELLNEKVEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTRNLSDLIPLNTVWS
GNPQAPCPFYPPGSPPLMQVATGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGRSMLPLTLAAGGDYYDVGGDEDTSLSFCPLAELSSDADRAWASEWIESLVAMQGITITPDHRNAIS
RQIGLMAVAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAETDGLALGLFQAFEIEQVMSMGERN
LVPVLTYLFRRIEKRLTGAPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAERSGI
IDVLKESCPTKICLPNGAAREPGTREFYERIGFNQRQIQIVASAIPKREYYVASPDGRRLFDMALGPLTL
SFVGASSKDDLKRIRALHEEHGPEWPLHWLQQRGIKDAASLLNDT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15414 | GenBank | WP_245461678 |
| Name | traG_U8P69_RS24925_pRlX1 |
UniProt ID | _ |
| Length | 667 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 667 a.a. Molecular weight: 71799.22 Da Isoelectric Point: 9.1969
>WP_245461678.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVVGPAALMILAVIGMTGIEQWLSGIGKTEAARLAWGRARIALPYAASAAIGVLFLFACAGSI
NIKRAGWGVLAGCIGTMLIAAVRETLRLSALMKTLPAGKTVLAYLDPATSIGASAALLCAGFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDIVGSQAFRADSAENWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSDHTDE
KDQTLRRVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQLNGRALFLLDEVARLGY
MRILETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETAEYISKRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIIFTAGNAPLRCARAIWFRRADMRACVG
ENRFYSMGGGEREGSNSTPAVNPYETKPTVTSAGEEG
MTINRIALVVGPAALMILAVIGMTGIEQWLSGIGKTEAARLAWGRARIALPYAASAAIGVLFLFACAGSI
NIKRAGWGVLAGCIGTMLIAAVRETLRLSALMKTLPAGKTVLAYLDPATSIGASAALLCAGFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDIVGSQAFRADSAENWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSDHTDE
KDQTLRRVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQLNGRALFLLDEVARLGY
MRILETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETAEYISKRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIIFTAGNAPLRCARAIWFRRADMRACVG
ENRFYSMGGGEREGSNSTPAVNPYETKPTVTSAGEEG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 151583..161052
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| U8P69_RS24735 (U8P69_24725) | 146829..148094 | - | 1266 | WP_327206240 | plasmid replication protein RepC | - |
| U8P69_RS24740 (U8P69_24730) | 148333..149373 | - | 1041 | WP_128411487 | plasmid partitioning protein RepB | - |
| U8P69_RS24745 (U8P69_24735) | 149358..150575 | - | 1218 | WP_128401849 | plasmid partitioning protein RepA | - |
| U8P69_RS24750 (U8P69_24740) | 150939..151568 | + | 630 | WP_128401848 | acyl-homoserine-lactone synthase TraI | - |
| U8P69_RS24755 (U8P69_24745) | 151583..152548 | + | 966 | WP_327206241 | P-type conjugative transfer ATPase TrbB | virB11 |
| U8P69_RS24760 (U8P69_24750) | 152538..152912 | + | 375 | WP_128401846 | TrbC/VirB2 family protein | virB2 |
| U8P69_RS24765 (U8P69_24755) | 152909..153208 | + | 300 | WP_128401845 | conjugal transfer protein TrbD | virB3 |
| U8P69_RS24770 (U8P69_24760) | 153218..155665 | + | 2448 | WP_128411486 | conjugal transfer protein TrbE | virb4 |
| U8P69_RS24775 (U8P69_24765) | 155637..156437 | + | 801 | WP_128401843 | P-type conjugative transfer protein TrbJ | virB5 |
| U8P69_RS24780 (U8P69_24770) | 156625..157800 | + | 1176 | WP_128401842 | P-type conjugative transfer protein TrbL | virB6 |
| U8P69_RS24785 (U8P69_24775) | 157820..158482 | + | 663 | WP_128401841 | conjugal transfer protein TrbF | virB8 |
| U8P69_RS24790 (U8P69_24780) | 158497..159321 | + | 825 | WP_128401840 | P-type conjugative transfer protein TrbG | virB9 |
| U8P69_RS24795 (U8P69_24785) | 159353..159751 | + | 399 | WP_327206242 | conjugal transfer protein TrbH | - |
| U8P69_RS24800 (U8P69_24790) | 159763..161052 | + | 1290 | WP_128401838 | IncP-type conjugal transfer protein TrbI | virB10 |
| U8P69_RS24805 (U8P69_24795) | 161637..162431 | - | 795 | WP_245461681 | autoinducer binding domain-containing protein | - |
| U8P69_RS24810 (U8P69_24800) | 162713..163444 | - | 732 | WP_128401837 | response regulator transcription factor | - |
| U8P69_RS24815 (U8P69_24805) | 164480..164776 | + | 297 | WP_245462682 | winged helix-turn-helix domain-containing protein | - |
Host bacterium
| ID | 21253 | GenBank | NZ_CP140852 |
| Plasmid name | pRlX1 | Incompatibility group | - |
| Plasmid size | 655862 bp | Coordinate of oriT [Strand] | 183925..183962 [-] |
| Host baterium | Rhizobium beringeri strain TP24 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | hupE2 |
| Degradation gene | - |
| Symbiosis gene | noeL |
| Anti-CRISPR | AcrIIA7 |