Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120672 |
| Name | oriT_pBSKP1 |
| Organism | Klebsiella variicola subsp. tropica strain BSK_177V2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP113790 (128952..129000 [+], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pBSKP1
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file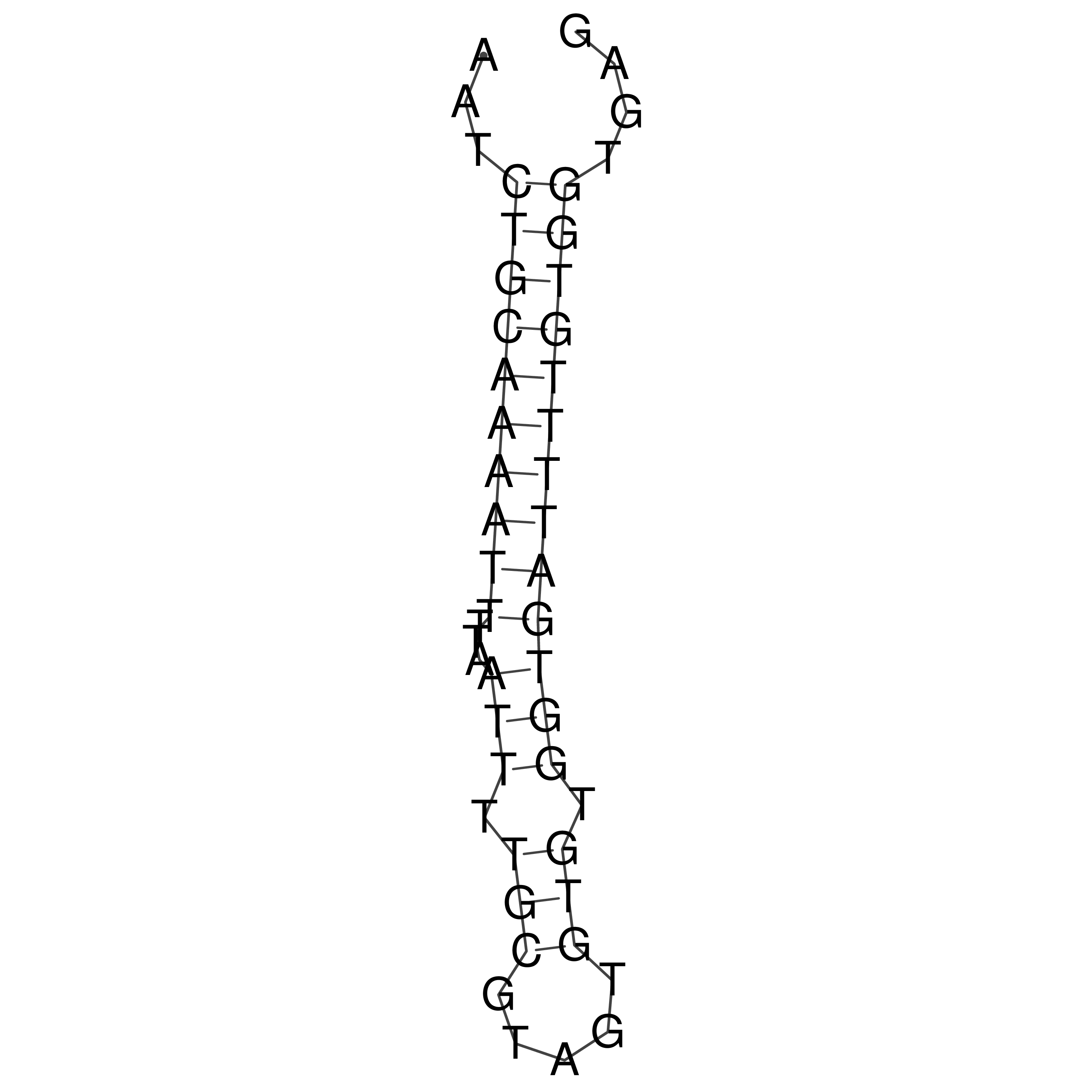
Relaxase
| ID | 13028 | GenBank | WP_040212835 |
| Name | traI_OUI59_RS28065_pBSKP1 |
UniProt ID | A0A509A0S3 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190231.91 Da Isoelectric Point: 6.2218
>WP_040212835.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella/Raoultella group]
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINTTQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARAGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTIQ
AQIRSGQLLNVPVAHGHGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPAQEKAIQK
ALSEGETLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINTTQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARAGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTIQ
AQIRSGQLLNVPVAHGHGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPAQEKAIQK
ALSEGETLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7287 | GenBank | WP_020805752 |
| Name | WP_020805752_pBSKP1 |
UniProt ID | A0A377TIM3 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 14772.81 Da Isoelectric Point: 4.5715
>WP_020805752.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Gammaproteobacteria]
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A377TIM3 |
T4CP
| ID | 15294 | GenBank | WP_023313941 |
| Name | traD_OUI59_RS28070_pBSKP1 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85666.78 Da Isoelectric Point: 5.2499
>WP_023313941.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGTADTNSHAGEQPEPVSQPA
PAEVTVSPAPVKAPATTKMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNKDGVIEDMQAYDAWADEQTLRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGTADTNSHAGEQPEPVSQPA
PAEVTVSPAPVKAPATTKMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNKDGVIEDMQAYDAWADEQTLRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 119829..129552
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| OUI59_RS28070 (OUI59_28070) | 119829..122138 | - | 2310 | WP_023313941 | type IV conjugative transfer system coupling protein TraD | virb4 |
| OUI59_RS28075 (OUI59_28075) | 122267..122803 | - | 537 | WP_268657994 | hypothetical protein | - |
| OUI59_RS28080 (OUI59_28080) | 122818..122919 | - | 102 | Protein_126 | type IV conjugative transfer system protein TraV | - |
| OUI59_RS28085 (OUI59_28085) | 122942..123538 | - | 597 | WP_023316398 | conjugal transfer pilus-stabilizing protein TraP | - |
| OUI59_RS28090 (OUI59_28090) | 123531..124955 | - | 1425 | WP_023316397 | F-type conjugal transfer pilus assembly protein TraB | traB |
| OUI59_RS28095 (OUI59_28095) | 124955..125695 | - | 741 | WP_020804840 | type-F conjugative transfer system secretin TraK | traK |
| OUI59_RS28100 (OUI59_28100) | 125682..126248 | - | 567 | WP_020316627 | type IV conjugative transfer system protein TraE | traE |
| OUI59_RS28105 (OUI59_28105) | 126268..126573 | - | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| OUI59_RS28110 (OUI59_28110) | 126587..126955 | - | 369 | WP_020316649 | type IV conjugative transfer system pilin TraA | - |
| OUI59_RS28115 (OUI59_28115) | 127024..127224 | - | 201 | WP_048289787 | TraY domain-containing protein | - |
| OUI59_RS28120 (OUI59_28120) | 127310..128011 | - | 702 | WP_023316396 | hypothetical protein | - |
| OUI59_RS28125 (OUI59_28125) | 128249..128641 | - | 393 | WP_020805752 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| OUI59_RS28130 (OUI59_28130) | 129145..129552 | + | 408 | WP_279615448 | transglycosylase SLT domain-containing protein | virB1 |
| OUI59_RS28135 (OUI59_28135) | 129590..130120 | - | 531 | WP_020277941 | antirestriction protein | - |
| OUI59_RS28140 (OUI59_28140) | 130745..131578 | - | 834 | WP_020277940 | N-6 DNA methylase | - |
| OUI59_RS28145 (OUI59_28145) | 131625..131855 | - | 231 | WP_020805755 | hypothetical protein | - |
| OUI59_RS28150 (OUI59_28150) | 131943..132290 | - | 348 | WP_020316654 | hypothetical protein | - |
| OUI59_RS28155 (OUI59_28155) | 132356..132736 | - | 381 | WP_015632488 | hypothetical protein | - |
| OUI59_RS28160 (OUI59_28160) | 133253..133603 | - | 351 | WP_020277938 | hypothetical protein | - |
| OUI59_RS28165 (OUI59_28165) | 133600..133872 | - | 273 | WP_023280873 | hypothetical protein | - |
| OUI59_RS28170 (OUI59_28170) | 134062..134547 | + | 486 | WP_020277937 | hypothetical protein | - |
Host bacterium
| ID | 21101 | GenBank | NZ_CP113790 |
| Plasmid name | pBSKP1 | Incompatibility group | IncFIB |
| Plasmid size | 154584 bp | Coordinate of oriT [Strand] | 128952..129000 [+] |
| Host baterium | Klebsiella variicola subsp. tropica strain BSK_177V2 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsC, arsB, arsR, arsH, silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, fecD, fecE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |