Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120531 |
| Name | oriT_p01A15005_B_KPC |
| Organism | Klebsiella oxytoca strain 01A15CRGN005 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_OQ821011 (46697..46745 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file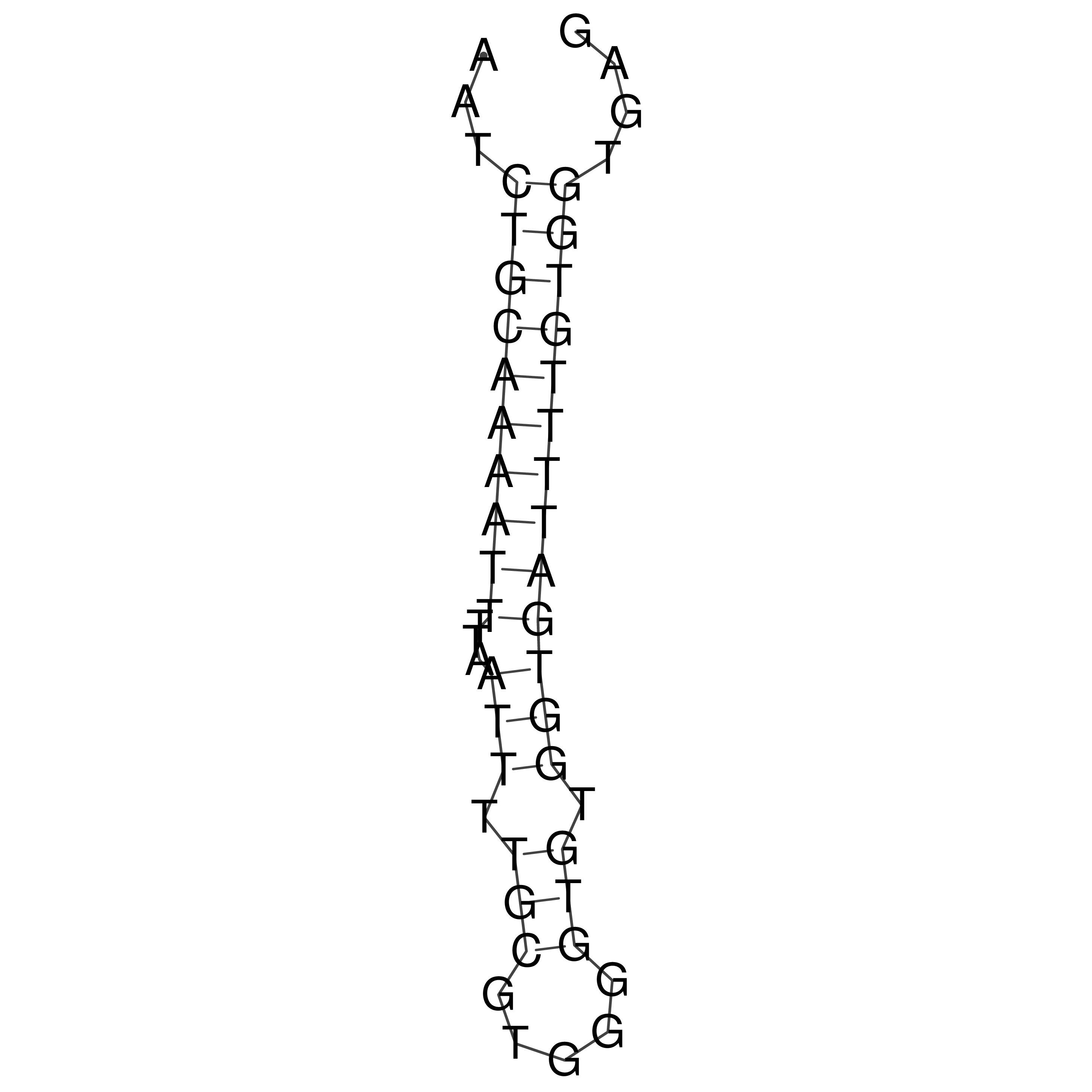
Relaxase
| ID | 12931 | GenBank | WP_319381996 |
| Name | traI_SLH58_RS00510_p01A15005_B_KPC |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190170.84 Da Isoelectric Point: 6.2774
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMEERWQGKGAEALGLDGKAVDKAVFTELLKGKLPDGSDLTRM
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLNDAHNQAVTVAVKQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQTAFGKLYREALKPLVE
KLGYETKVVGKHGMWEMKDVPVEPFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADAHAAELARAPAAPVKTDGPDIADVVTRAIAGLSDRKVQFTYADVLARTVGQLE
AKPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDAGVVREKP
FSDAVSVMAQDRPAMGIVAGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSREFLAGDARLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGKQATADIISEPDKSARYSRLAQEFAASVREGQESVAQISGTREQNVLNNVIRDTLKNEGALGNRD
MTVTALTPVWLDSKSRGVRDYYREGMVMERWDPEARKHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRTDKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQTLSVGSSPFDGI
KVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDSARTTEKLARHTAFSVVSE
QLKVRSGETDLEAAIAQQKAGLHTPAEQAIHLSVPLLESNSLTFTRPQLLATALETGGGKVPMKDIEATI
QAQIKAGSLLNVPVASGHGNDLLISRQTWDAEKSVLTRVLEGKDAVAPLMDRVPGSLMTDLTAGQRAATR
MILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLH
DTQLQQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDVSRALSTIEQVTPEQVPRKDGAWVPENSVTEFTPAQEKAIQ
DALKEGKEIPPGTPVSLHDAIVKDYTGRTPAAQASTLIITHLNADRRALNGLIHDARVQNGEVSKEEVTL
PVLVTSNIRDGELRKLSTWSEHQGAVALLDNTYQRIESVDKENQLITLKDNEGNIRLLSPREASAEGVTL
YRPEHITVSQGDRMRFSKSDPERGYVANSVWEVKAFSGDSVTLSDGKTTRTLNPKADEAQRHIDLAYAIT
AHGAQGASEPFAIALEGVEGRRKALVSFESAYVALSRMKQHAQVYTDNREGWTTAVSRSQEKASAHDVLE
PQNARAVRNAGQLYGRAQPLDETAAGRAALQQSGLARGRSPGKFISPGKKYPQPHVALPAFDKNGKEAGI
WLSPLTDRDGRLEAIGGEGRIMGDETARFVALQSSRNGESLLAGNMGEGVRIARDNPDSGVIVRLAGDDR
PWNPEAITGGRIWADPLPNPPLPDTGTDIPLPPEVLAQRAAEEAQRREMEKQAEQATREVAGEEKKAGEP
GERIKEVIGDVIRGLERDRPGEEKTRLPDDPQTRRQEAAIQQVASESLQRERLQQMERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7266 | GenBank | WP_032426796 |
| Name | WP_032426796_p01A15005_B_KPC |
UniProt ID | _ |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 15036.10 Da Isoelectric Point: 4.5326
MGKVQAYPSDEVCEKINAIVEKRRMEGAKEKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMRTFFPDEQEEADE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15187 | GenBank | WP_319381995 |
| Name | traD_SLH58_RS00505_p01A15005_B_KPC |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 85519.29 Da Isoelectric Point: 4.8808
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRTLDTRVDARLSALLEAREAEGSLARALFTPDAPEPEQSDADSKPSPGQPEPESAP
VSPAPVKSASTAETLAAQPSVKASEAPQAKVTTVPLTRPAPAATPAAASATASSAAATAAATGGTEQALA
QQPADTGQEMMPAGMNEDGEIEDMQAYDAWLADEQTQREMQRREEVNINHSHRRDEQDDIEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 15188 | GenBank | WP_004152620 |
| Name | traD_SLH58_RS00595_p01A15005_B_KPC |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85871.84 Da Isoelectric Point: 5.0101
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 49163..72212
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SLH58_RS00340 | 45202..45378 | - | 177 | WP_319382015 | hypothetical protein | - |
| SLH58_RS00345 (KEGJGPDJ_00059) | 45565..46107 | + | 543 | WP_319382016 | antirestriction protein | - |
| SLH58_RS00350 (KEGJGPDJ_00060) | 46130..46552 | - | 423 | WP_239605204 | transglycosylase SLT domain-containing protein | - |
| SLH58_RS00355 (KEGJGPDJ_00061) | 47058..47450 | + | 393 | WP_032426796 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| SLH58_RS00360 (KEGJGPDJ_00062) | 47688..48380 | + | 693 | WP_032426795 | conjugal transfer protein TrbJ | - |
| SLH58_RS00365 | 48556..48720 | + | 165 | WP_032426810 | TraY domain-containing protein | - |
| SLH58_RS00370 (KEGJGPDJ_00063) | 48784..49149 | + | 366 | WP_064381359 | type IV conjugative transfer system pilin TraA | - |
| SLH58_RS00375 (KEGJGPDJ_00064) | 49163..49468 | + | 306 | WP_020323523 | type IV conjugative transfer system protein TraL | traL |
| SLH58_RS00380 (KEGJGPDJ_00065) | 49488..50054 | + | 567 | WP_064373744 | type IV conjugative transfer system protein TraE | traE |
| SLH58_RS00385 (KEGJGPDJ_00066) | 50041..50781 | + | 741 | WP_319382017 | type-F conjugative transfer system secretin TraK | traK |
| SLH58_RS00390 (KEGJGPDJ_00067) | 50781..52199 | + | 1419 | WP_319382018 | F-type conjugal transfer pilus assembly protein TraB | traB |
| SLH58_RS00395 (KEGJGPDJ_00068) | 52192..52788 | + | 597 | WP_319382019 | conjugal transfer protein TraP | - |
| SLH58_RS00400 | 52856..52993 | + | 138 | Protein_79 | conjugal transfer protein TraP | - |
| SLH58_RS00405 (KEGJGPDJ_00070) | 53016..53582 | + | 567 | WP_319382020 | type IV conjugative transfer system lipoprotein TraV | traV |
| SLH58_RS00410 (KEGJGPDJ_00071) | 53699..54103 | + | 405 | WP_319382021 | hypothetical protein | - |
| SLH58_RS00415 (KEGJGPDJ_00072) | 54132..54425 | + | 294 | WP_319382022 | hypothetical protein | - |
| SLH58_RS00420 (KEGJGPDJ_00073) | 54449..54673 | + | 225 | WP_064381372 | hypothetical protein | - |
| SLH58_RS00425 (KEGJGPDJ_00074) | 54670..55074 | + | 405 | WP_319382023 | hypothetical protein | - |
| SLH58_RS00430 (KEGJGPDJ_00075) | 55205..57844 | + | 2640 | WP_319382024 | type IV secretion system protein TraC | virb4 |
| SLH58_RS00435 (KEGJGPDJ_00076) | 57877..58278 | + | 402 | WP_319382034 | type-F conjugative transfer system protein TrbI | - |
| SLH58_RS00440 (KEGJGPDJ_00077) | 58275..58901 | + | 627 | WP_319382025 | type-F conjugative transfer system protein TraW | traW |
| SLH58_RS00445 (KEGJGPDJ_00078) | 58921..59904 | + | 984 | WP_319382035 | conjugal transfer pilus assembly protein TraU | traU |
| SLH58_RS00450 (KEGJGPDJ_00079) | 59917..60534 | + | 618 | WP_319382026 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| SLH58_RS00455 (KEGJGPDJ_00080) | 60531..62486 | + | 1956 | WP_319382027 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| SLH58_RS00460 (KEGJGPDJ_00081) | 62524..62805 | + | 282 | WP_319382028 | hypothetical protein | - |
| SLH58_RS00465 (KEGJGPDJ_00082) | 62795..63022 | + | 228 | WP_064174417 | conjugal transfer protein TrbE | - |
| SLH58_RS00470 (KEGJGPDJ_00083) | 63033..63359 | + | 327 | WP_135800727 | hypothetical protein | - |
| SLH58_RS00475 (KEGJGPDJ_00084) | 63378..64130 | + | 753 | WP_319382029 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| SLH58_RS00480 (KEGJGPDJ_00085) | 64141..64380 | + | 240 | WP_319382030 | type-F conjugative transfer system pilin chaperone TraQ | - |
| SLH58_RS00485 (KEGJGPDJ_00086) | 64352..64927 | + | 576 | WP_319382031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| SLH58_RS00490 (KEGJGPDJ_00087) | 64914..66284 | + | 1371 | WP_319382032 | conjugal transfer pilus assembly protein TraH | traH |
| SLH58_RS00495 (KEGJGPDJ_00088) | 66284..69127 | + | 2844 | WP_319382033 | conjugal transfer mating-pair stabilization protein TraG | traG |
| SLH58_RS00500 (KEGJGPDJ_00089) | 69138..69674 | + | 537 | WP_301718420 | hypothetical protein | - |
| SLH58_RS00505 (KEGJGPDJ_00090) | 69912..72212 | + | 2301 | WP_319381995 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 20960 | GenBank | NZ_OQ821011 |
| Plasmid name | p01A15005_B_KPC | Incompatibility group | IncR |
| Plasmid size | 134488 bp | Coordinate of oriT [Strand] | 46697..46745 [-] |
| Host baterium | Klebsiella oxytoca strain 01A15CRGN005 |
Cargo genes
| Drug resistance gene | blaKPC-2, blaOXA-9, blaTEM-1A, aph(6)-Id, aph(3'')-Ib |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |