Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120466 |
| Name | oriT_pEFM61-3 |
| Organism | Enterococcus faecalis strain M61 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP113835 (28131..28166 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 1..8, 13..20 (GTAACATG..CATGTTAC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pEFM61-3
GTAACATGCTATCATGTTACTATGCTCGCGAAAGGT
GTAACATGCTATCATGTTACTATGCTCGCGAAAGGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file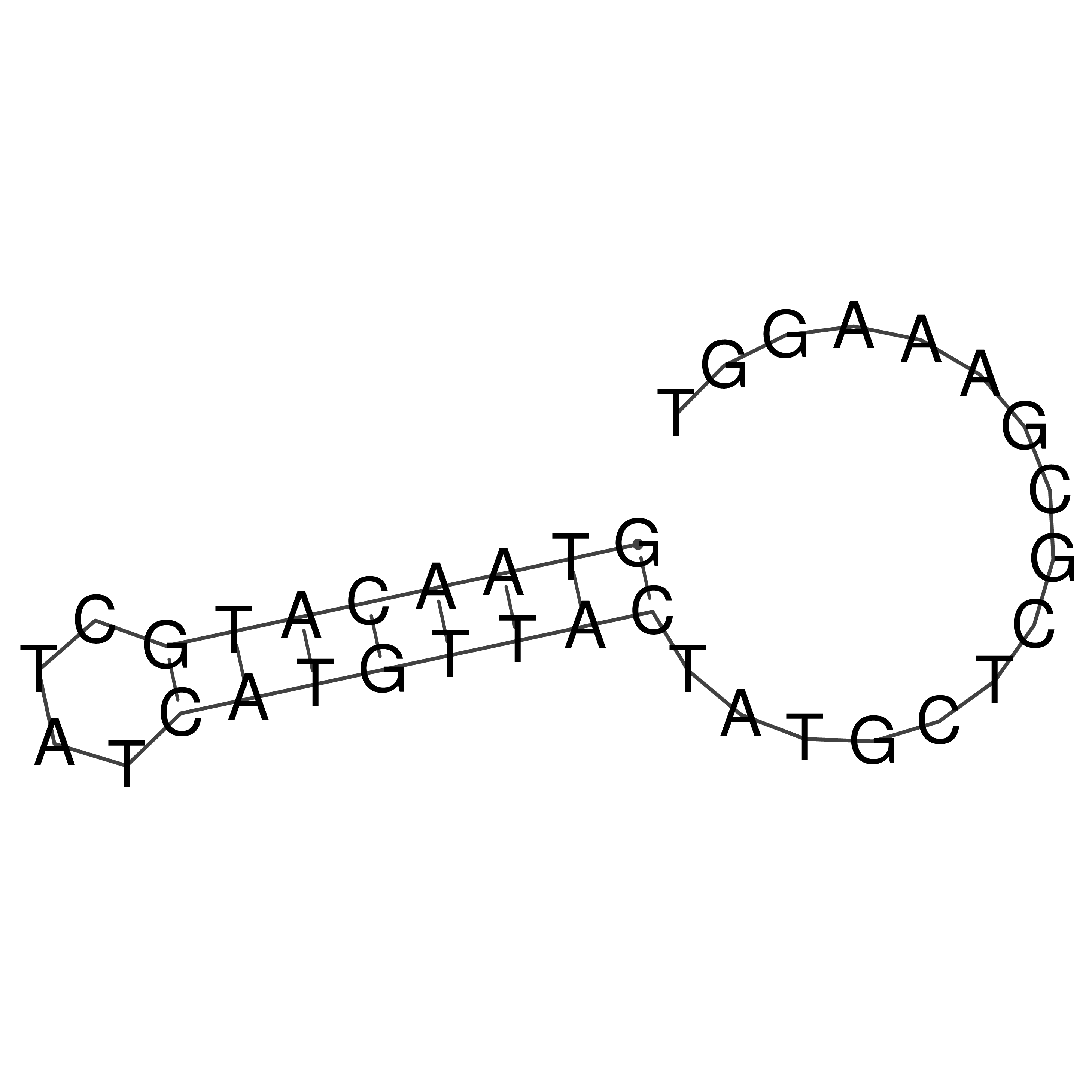
Relaxase
| ID | 12888 | GenBank | WP_104670895 |
| Name | Relaxase_OZF29_RS14375_pEFM61-3 |
UniProt ID | _ |
| Length | 571 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 571 a.a. Molecular weight: 66176.96 Da Isoelectric Point: 6.1074
>WP_104670895.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterococcus]
MVYTKHFTIHTMEHLNQSKEYVQKASKTTVDKSSSGNTHYEQLFPYIMNADKTLSKQLVSGHGITDVTNA
ANEFLYTKERVAKQKGTYLMFDPETETMVFDKAAIEKGNGRGQAVLGHHLIQSFSPEDDLTPEEIHEIGR
QTVLEFTGGEHEFVIATHVDRKHIHNHIIINSTNLFTGKALPWKVVTFENGETKDYTKELFEKVSDKISA
KYGAKIIEKSPKNSHKKYTMWQTESIYKSKIKSRLDFLLDHSSSIEDFKCKAEALELHVDFSGKWATYRL
LDEPQIKNTRSRSLNKKNPEKYNLKQIEERLKENTASFSIEEVVDRYEEKINVVKNDFDYQLIVEPWQIS
HKTPKGYYLNVDFGIENHGQIFLGAYKVDELEDGNYSLFVKKNDFFYFMNEKNGQRNRYMTGYTLAKQLS
LYNGTVPLKKEPVMSTIDEIVDAINFLAEHDVIEGNQMERLEKQLVEVFKTAEESLETLDEKILQLNQVG
KLLLANELENNKENLPILKQLEEIEELTYEDVKNELTSLKLSRKLLQEKMETTVEKINELHGVQTVVERK
TKEENLGKIEK
MVYTKHFTIHTMEHLNQSKEYVQKASKTTVDKSSSGNTHYEQLFPYIMNADKTLSKQLVSGHGITDVTNA
ANEFLYTKERVAKQKGTYLMFDPETETMVFDKAAIEKGNGRGQAVLGHHLIQSFSPEDDLTPEEIHEIGR
QTVLEFTGGEHEFVIATHVDRKHIHNHIIINSTNLFTGKALPWKVVTFENGETKDYTKELFEKVSDKISA
KYGAKIIEKSPKNSHKKYTMWQTESIYKSKIKSRLDFLLDHSSSIEDFKCKAEALELHVDFSGKWATYRL
LDEPQIKNTRSRSLNKKNPEKYNLKQIEERLKENTASFSIEEVVDRYEEKINVVKNDFDYQLIVEPWQIS
HKTPKGYYLNVDFGIENHGQIFLGAYKVDELEDGNYSLFVKKNDFFYFMNEKNGQRNRYMTGYTLAKQLS
LYNGTVPLKKEPVMSTIDEIVDAINFLAEHDVIEGNQMERLEKQLVEVFKTAEESLETLDEKILQLNQVG
KLLLANELENNKENLPILKQLEEIEELTYEDVKNELTSLKLSRKLLQEKMETTVEKINELHGVQTVVERK
TKEENLGKIEK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15137 | GenBank | WP_104670887 |
| Name | t4cp2_OZF29_RS14335_pEFM61-3 |
UniProt ID | _ |
| Length | 603 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 603 a.a. Molecular weight: 69893.94 Da Isoelectric Point: 6.5211
>WP_104670887.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Enterococcus]
MQVYKKEFKPYLMLAICLFFVGYALGNFFWLLPGYDYFSKMNYLFTKDLLSYANENFFLFFFTPSFLAFG
TGFLGFLIGLLAYVRDNDRGIYRNGEEYGSARFATDEEMAKFSDKDPKNNMIISKKVQIGLFNFRLPIKI
QKNKNVMVVGDSGSAKTLAYILTNILQLNASFVVTDPDGGIVHKVGKLLKKFGYKIKILDLNQLVNSDTF
NVFQYIRTELDIDRVLEAITEGTKKTEQQGEDFWIQAEALLIRSFIAYLWFDGQDNDYMPNLSMIADMLR
HVERKDKKVPSPVEQWFEEQNQLRPNNYAYKQWTLYNDSYKAETRASVLAIASARYSVFDHEQVADMIRK
DTMEIDKWNEEKTAVFIAIPETNKSYNFIAAIFMATVMETLRHKSDQVRLGKLQLPKEKKLLHVRFLIDE
FANIGRIPNFEEALATFRKREMSFSIILQSLAQLQKMYKYGWEGIVNNCSTLLFLGGDEEKTTKYLSHRA
DKQTISLRKYSKSHGRNGGGSESRDRTGRDLLTPGEIGHLDGDECLVYISKEYVYKDKKYYAFDHPLGSM
MGDEPGDENWYTYKRYLTEEEEILDKVRPENIIDHGVIGDEVA
MQVYKKEFKPYLMLAICLFFVGYALGNFFWLLPGYDYFSKMNYLFTKDLLSYANENFFLFFFTPSFLAFG
TGFLGFLIGLLAYVRDNDRGIYRNGEEYGSARFATDEEMAKFSDKDPKNNMIISKKVQIGLFNFRLPIKI
QKNKNVMVVGDSGSAKTLAYILTNILQLNASFVVTDPDGGIVHKVGKLLKKFGYKIKILDLNQLVNSDTF
NVFQYIRTELDIDRVLEAITEGTKKTEQQGEDFWIQAEALLIRSFIAYLWFDGQDNDYMPNLSMIADMLR
HVERKDKKVPSPVEQWFEEQNQLRPNNYAYKQWTLYNDSYKAETRASVLAIASARYSVFDHEQVADMIRK
DTMEIDKWNEEKTAVFIAIPETNKSYNFIAAIFMATVMETLRHKSDQVRLGKLQLPKEKKLLHVRFLIDE
FANIGRIPNFEEALATFRKREMSFSIILQSLAQLQKMYKYGWEGIVNNCSTLLFLGGDEEKTTKYLSHRA
DKQTISLRKYSKSHGRNGGGSESRDRTGRDLLTPGEIGHLDGDECLVYISKEYVYKDKKYYAFDHPLGSM
MGDEPGDENWYTYKRYLTEEEEILDKVRPENIIDHGVIGDEVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3839..19451
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| OZF29_RS14245 (OZF29_14245) | 2463..2741 | + | 279 | WP_104670904 | hypothetical protein | - |
| OZF29_RS14255 (OZF29_14255) | 3200..3523 | + | 324 | WP_002383600 | DUF5406 family protein | - |
| OZF29_RS14260 (OZF29_14260) | 3839..7717 | + | 3879 | WP_104670964 | LPXTG-anchored aggregation substance | prgB |
| OZF29_RS14265 (OZF29_14265) | 8057..8197 | + | 141 | WP_104670876 | LPXTG cell wall anchor domain-containing protein | prgC |
| OZF29_RS14270 (OZF29_14270) | 8261..8617 | + | 357 | WP_010711024 | pheromone response system RNA-binding regulator PrgU | - |
| OZF29_RS14275 (OZF29_14275) | 8645..9499 | + | 855 | WP_104670877 | pheromone response LPXTG-anchored adhesin PrgC | prgC |
| OZF29_RS14280 (OZF29_14280) | 9817..10713 | + | 897 | WP_104670878 | type III secretion system protein PrgD | - |
| OZF29_RS14285 (OZF29_14285) | 10732..11166 | + | 435 | WP_104670879 | type III secretion system protein PrgE | - |
| OZF29_RS14290 (OZF29_14290) | 11191..11418 | + | 228 | WP_104670880 | type III secretion system protein PrgF | - |
| OZF29_RS14295 (OZF29_14295) | 11431..12222 | + | 792 | WP_104670881 | type III secretion system protein PrgH | prgHb |
| OZF29_RS14300 (OZF29_14300) | 12224..12577 | + | 354 | WP_104670882 | PrgI family mobile element protein | prgIc |
| OZF29_RS14305 (OZF29_14305) | 12531..14918 | + | 2388 | WP_161975846 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| OZF29_RS14310 (OZF29_14310) | 14930..17545 | + | 2616 | WP_104670883 | phage tail tip lysozyme | prgK |
| OZF29_RS14315 (OZF29_14315) | 17585..18199 | + | 615 | WP_104670884 | hypothetical protein | prgL |
| OZF29_RS14320 (OZF29_14320) | 18192..18422 | + | 231 | WP_010711013 | hypothetical protein | - |
| OZF29_RS14325 (OZF29_14325) | 18445..18924 | + | 480 | WP_104670885 | hypothetical protein | - |
| OZF29_RS14330 (OZF29_14330) | 18966..19451 | + | 486 | WP_161975848 | DUF3801 domain-containing protein | gbs1365 |
| OZF29_RS14335 (OZF29_14335) | 19451..21262 | + | 1812 | WP_104670887 | VirD4-like conjugal transfer protein, CD1115 family | - |
| OZF29_RS14340 (OZF29_14340) | 21347..21910 | + | 564 | WP_104670888 | hypothetical protein | - |
| OZF29_RS14345 (OZF29_14345) | 21948..22421 | + | 474 | WP_104670889 | single-stranded DNA-binding protein | - |
| OZF29_RS14350 (OZF29_14350) | 22511..22711 | + | 201 | WP_104670890 | hypothetical protein | - |
Host bacterium
| ID | 20895 | GenBank | NZ_CP113835 |
| Plasmid name | pEFM61-3 | Incompatibility group | - |
| Plasmid size | 49275 bp | Coordinate of oriT [Strand] | 28131..28166 [+] |
| Host baterium | Enterococcus faecalis strain M61 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | prgB/asc10, asa1 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |