Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120461 |
| Name | oriT_AR_0116|unnamed3 |
| Organism | Citrobacter freundii strain AR_0116 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP032181 (69973..70021 [+], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_AR_0116|unnamed3
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file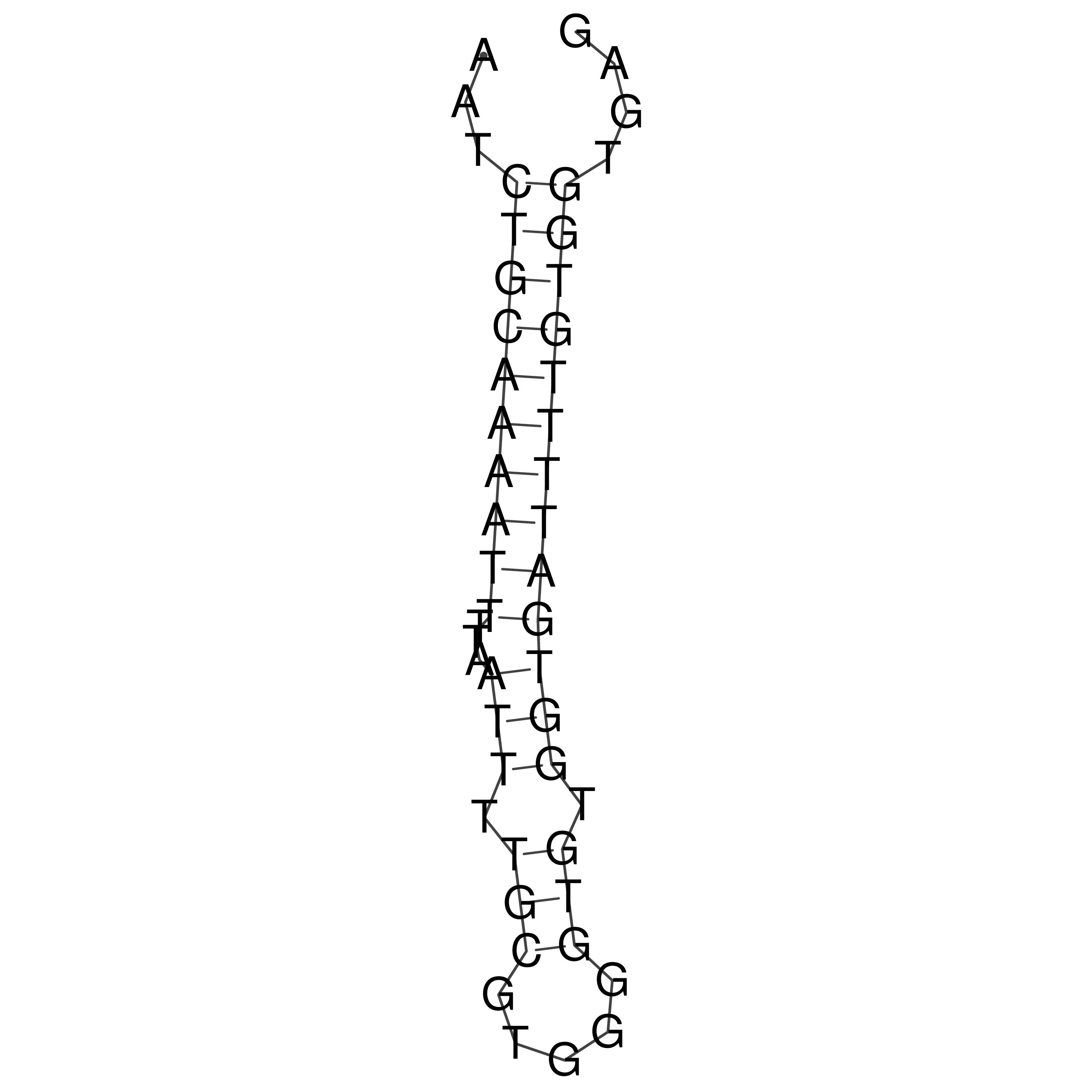
Relaxase
| ID | 12885 | GenBank | WP_119173777 |
| Name | traI_AM363_RS02515_AR_0116|unnamed3 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190683.66 Da Isoelectric Point: 6.2410
>WP_119173777.1 conjugative transfer relaxase/helicase TraI [Citrobacter freundii]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGTNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKLYREALKPLVE
KLGYETDVVGKHGMWEMKDVPVEPFSTRTREVREAAGPDASLKSRDVAALDTRKSKETLDPAEKVVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVKADGPDIGDVVTRAIAGLSDRKVQFTYADVLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLILKDREGDRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQLRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLTQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPLAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGTNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKLYREALKPLVE
KLGYETDVVGKHGMWEMKDVPVEPFSTRTREVREAAGPDASLKSRDVAALDTRKSKETLDPAEKVVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVKADGPDIGDVVTRAIAGLSDRKVQFTYADVLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLILKDREGDRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQLRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLTQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPLAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7259 | GenBank | WP_022644939 |
| Name | WP_022644939_AR_0116|unnamed3 |
UniProt ID | W8QGP1 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 15048.16 Da Isoelectric Point: 4.5248
>WP_022644939.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacteriaceae]
MAKVQAYVSDEVAEKINAIVEKRRVEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNKAL
LEYVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMLRFFPDEDNEVEE
MAKVQAYVSDEVAEKINAIVEKRRVEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNKAL
LEYVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMLRFFPDEDNEVEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W8QGP1 |
T4CP
| ID | 15134 | GenBank | WP_119173778 |
| Name | traD_AM363_RS02520_AR_0116|unnamed3 |
UniProt ID | _ |
| Length | 773 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 773 a.a. Molecular weight: 86126.05 Da Isoelectric Point: 5.1241
>WP_119173778.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKARPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQHEPVKQSE
PAGDPVSPVPVKAPSAAKTSVAAEPVKSPETPQPVVPATRVTTVPLIRTKPASAANGTAVATGGATAHSG
GTEQELKPQPADTGQEMMPAGMNEDGEIEDMAAYDAWLAGEQTQREMQRREEVNINHSHRHDEQDDVEIG
GNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKARPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQHEPVKQSE
PAGDPVSPVPVKAPSAAKTSVAAEPVKSPETPQPVVPATRVTTVPLIRTKPASAANGTAVATGGATAHSG
GTEQELKPQPADTGQEMMPAGMNEDGEIEDMAAYDAWLAGEQTQREMQRREEVNINHSHRHDEQDDVEIG
GNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 40061..67596
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AM363_RS02520 (AM363_02530) | 40061..42382 | - | 2322 | WP_119173778 | type IV conjugative transfer system coupling protein TraD | virb4 |
| AM363_RS02525 (AM363_02535) | 42454..42717 | - | 264 | WP_162906117 | hypothetical protein | - |
| AM363_RS29685 | 42738..43430 | - | 693 | WP_162762153 | hypothetical protein | - |
| AM363_RS02535 (AM363_02545) | 43677..44348 | - | 672 | WP_088883382 | hypothetical protein | - |
| AM363_RS02540 (AM363_02550) | 44365..47190 | - | 2826 | WP_119173780 | conjugal transfer mating-pair stabilization protein TraG | traG |
| AM363_RS02545 (AM363_02555) | 47190..48560 | - | 1371 | WP_004159274 | conjugal transfer pilus assembly protein TraH | traH |
| AM363_RS02550 (AM363_02560) | 48547..48975 | - | 429 | WP_015065560 | hypothetical protein | - |
| AM363_RS02555 (AM363_02565) | 48968..49540 | - | 573 | WP_023292147 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| AM363_RS02560 (AM363_02570) | 49512..49751 | - | 240 | WP_004152687 | type-F conjugative transfer system pilin chaperone TraQ | - |
| AM363_RS02565 (AM363_02575) | 49762..50514 | - | 753 | WP_117072077 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| AM363_RS02570 (AM363_02580) | 50535..50861 | - | 327 | WP_119173781 | hypothetical protein | - |
| AM363_RS02575 (AM363_02585) | 50872..51099 | - | 228 | WP_012540032 | conjugal transfer protein TrbE | - |
| AM363_RS02580 (AM363_02590) | 51089..51370 | - | 282 | WP_012540038 | hypothetical protein | - |
| AM363_RS02585 (AM363_02595) | 51403..53358 | - | 1956 | WP_060598862 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| AM363_RS02590 (AM363_02600) | 53417..54064 | - | 648 | WP_039698503 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| AM363_RS02595 (AM363_02605) | 54209..54811 | - | 603 | WP_022631520 | hypothetical protein | - |
| AM363_RS02605 (AM363_02615) | 55532..56080 | - | 549 | WP_022631518 | hypothetical protein | - |
| AM363_RS02610 (AM363_02620) | 56095..57054 | - | 960 | WP_029497356 | conjugal transfer pilus assembly protein TraU | traU |
| AM363_RS02615 (AM363_02625) | 57098..57724 | - | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| AM363_RS02620 (AM363_02630) | 57724..58113 | - | 390 | WP_060446931 | type-F conjugative transfer system protein TrbI | - |
| AM363_RS02625 (AM363_02635) | 58113..60752 | - | 2640 | WP_020323518 | type IV secretion system protein TraC | virb4 |
| AM363_RS02630 (AM363_02640) | 60824..61222 | - | 399 | WP_162906118 | hypothetical protein | - |
| AM363_RS02635 (AM363_02645) | 61230..61520 | - | 291 | WP_020323516 | hypothetical protein | - |
| AM363_RS02640 (AM363_02650) | 61517..61921 | - | 405 | WP_049246239 | hypothetical protein | - |
| AM363_RS02645 (AM363_02655) | 61988..62299 | - | 312 | WP_100698965 | hypothetical protein | - |
| AM363_RS02650 (AM363_02660) | 62300..62518 | - | 219 | WP_119173783 | hypothetical protein | - |
| AM363_RS02655 (AM363_02665) | 62542..62826 | - | 285 | WP_119173789 | hypothetical protein | - |
| AM363_RS02660 (AM363_02670) | 62831..63241 | - | 411 | WP_020805613 | hypothetical protein | - |
| AM363_RS02665 (AM363_02675) | 63373..63942 | - | 570 | WP_023316399 | type IV conjugative transfer system lipoprotein TraV | traV |
| AM363_RS02670 (AM363_02680) | 63965..64561 | - | 597 | WP_023316398 | conjugal transfer pilus-stabilizing protein TraP | - |
| AM363_RS02675 (AM363_02685) | 64554..65978 | - | 1425 | WP_119173784 | F-type conjugal transfer pilus assembly protein TraB | traB |
| AM363_RS02680 (AM363_02690) | 65978..66718 | - | 741 | WP_119173785 | type-F conjugative transfer system secretin TraK | traK |
| AM363_RS02685 (AM363_02695) | 66705..67271 | - | 567 | WP_020316627 | type IV conjugative transfer system protein TraE | traE |
| AM363_RS02690 (AM363_02700) | 67291..67596 | - | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| AM363_RS02695 (AM363_02705) | 67610..67978 | - | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| AM363_RS02705 (AM363_02715) | 68332..69033 | - | 702 | WP_022644938 | hypothetical protein | - |
| AM363_RS02710 (AM363_02720) | 69268..69660 | - | 393 | WP_022644939 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| AM363_RS02715 (AM363_02725) | 70166..70573 | + | 408 | WP_109043189 | transglycosylase SLT domain-containing protein | - |
| AM363_RS02720 (AM363_02730) | 70611..71138 | - | 528 | WP_109043195 | antirestriction protein | - |
| AM363_RS02725 (AM363_02735) | 71551..71697 | - | 147 | WP_109043194 | succinate dehydrogenase flavoprotein subunit | - |
| AM363_RS02730 (AM363_02740) | 71790..72137 | - | 348 | WP_109043188 | hypothetical protein | - |
| AM363_RS02735 (AM363_02745) | 72204..72584 | - | 381 | WP_119173786 | hypothetical protein | - |
Host bacterium
| ID | 20890 | GenBank | NZ_CP032181 |
| Plasmid name | AR_0116|unnamed3 | Incompatibility group | - |
| Plasmid size | 81323 bp | Coordinate of oriT [Strand] | 69973..70021 [+] |
| Host baterium | Citrobacter freundii strain AR_0116 |
Cargo genes
| Drug resistance gene | blaKPC-2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |