Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120374 |
| Name | oriT_pRlX2 |
| Organism | Rhizobium beringeri strain TP6 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP140809 (243989..244029 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pRlX2
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file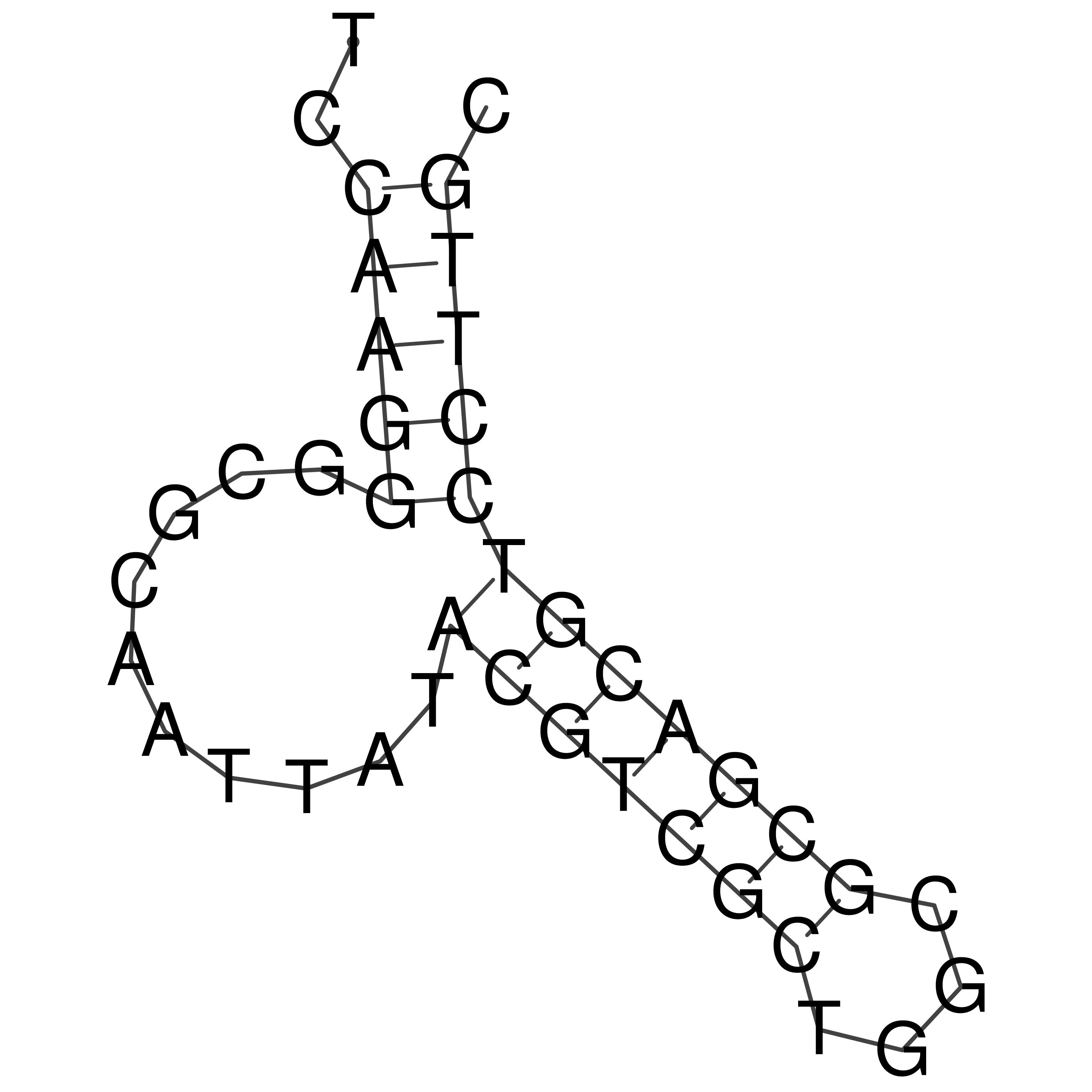
Relaxase
| ID | 12821 | GenBank | WP_130759159 |
| Name | traA_U8P73_RS30230_pRlX2 |
UniProt ID | _ |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 122958.22 Da Isoelectric Point: 9.6155
>WP_130759159.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADS
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDDKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVLLFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
VRLEAEMANRAIWLSGRASHGVREEVLQATFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRVGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAAGGIIAEIGEGEHRQVTIEQRFYNNLDHGYAT
TIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYEK
SALCRQAFRFAEARGLHLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGKALGLVRGSNKHSIPDTIK
EARPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAQPEAAFKAINVDAMVKDQSVAQST
LARIAGEPESFGALKGKTGVLASRGDKHDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKVS
VDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDGK
TFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADS
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDDKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVLLFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
VRLEAEMANRAIWLSGRASHGVREEVLQATFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRVGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAAGGIIAEIGEGEHRQVTIEQRFYNNLDHGYAT
TIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYEK
SALCRQAFRFAEARGLHLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGKALGLVRGSNKHSIPDTIK
EARPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAQPEAAFKAINVDAMVKDQSVAQST
LARIAGEPESFGALKGKTGVLASRGDKHDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKVS
VDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDGK
TFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15066 | GenBank | WP_130692298 |
| Name | traG_U8P73_RS30215_pRlX2 |
UniProt ID | _ |
| Length | 643 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 643 a.a. Molecular weight: 69483.71 Da Isoelectric Point: 9.1051
>WP_130692298.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSAFMTVPADKTVWAFLDPATSIGASAALLCACFALRVALIGN
AAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGGK
SPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLDP
RKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDEK
DQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAA
LVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGYM
RIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTV
EIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGT
NRFHMVGDTPRPA
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSAFMTVPADKTVWAFLDPATSIGASAALLCACFALRVALIGN
AAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGGK
SPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLDP
RKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDEK
DQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAA
LVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGYM
RIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTV
EIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGT
NRFHMVGDTPRPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 255676..263446
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| U8P73_RS30255 (U8P73_30250) | 250997..251371 | - | 375 | WP_130692288 | DUF5615 family PIN-like protein | - |
| U8P73_RS30260 (U8P73_30255) | 251368..252000 | - | 633 | WP_128408809 | DUF433 domain-containing protein | - |
| U8P73_RS30265 (U8P73_30260) | 252114..252356 | - | 243 | WP_128408808 | helix-turn-helix domain-containing protein | - |
| U8P73_RS30270 (U8P73_30265) | 252582..252923 | + | 342 | WP_130759157 | transcriptional repressor TraM | - |
| U8P73_RS30275 (U8P73_30270) | 252908..253612 | - | 705 | WP_128408806 | autoinducer binding domain-containing protein | - |
| U8P73_RS30280 (U8P73_30275) | 253921..255220 | - | 1300 | Protein_252 | IncP-type conjugal transfer protein TrbI | - |
| U8P73_RS30285 (U8P73_30280) | 255232..255672 | - | 441 | WP_130692284 | conjugal transfer protein TrbH | - |
| U8P73_RS30290 (U8P73_30285) | 255676..256488 | - | 813 | WP_130759156 | P-type conjugative transfer protein TrbG | virB9 |
| U8P73_RS30295 (U8P73_30290) | 256506..257168 | - | 663 | WP_326893021 | conjugal transfer protein TrbF | virB8 |
| U8P73_RS30300 (U8P73_30295) | 257189..258369 | - | 1181 | Protein_256 | P-type conjugative transfer protein TrbL | - |
| U8P73_RS30305 (U8P73_30300) | 258363..258563 | - | 201 | WP_130759154 | entry exclusion protein TrbK | - |
| U8P73_RS30310 (U8P73_30305) | 258560..259363 | - | 804 | WP_130759153 | P-type conjugative transfer protein TrbJ | virB5 |
| U8P73_RS30315 (U8P73_30310) | 259335..261793 | - | 2459 | Protein_259 | conjugal transfer protein TrbE | - |
| U8P73_RS30320 (U8P73_30315) | 261804..262103 | - | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| U8P73_RS30325 (U8P73_30320) | 262096..262479 | - | 384 | WP_063473648 | TrbC/VirB2 family protein | virB2 |
| U8P73_RS30330 (U8P73_30325) | 262469..263446 | - | 978 | WP_130692274 | P-type conjugative transfer ATPase TrbB | virB11 |
| U8P73_RS30335 (U8P73_30330) | 263443..264081 | - | 639 | WP_326893022 | acyl-homoserine-lactone synthase | - |
| U8P73_RS30340 (U8P73_30335) | 264559..264771 | + | 213 | WP_245458276 | hypothetical protein | - |
Host bacterium
| ID | 20803 | GenBank | NZ_CP140809 |
| Plasmid name | pRlX2 | Incompatibility group | - |
| Plasmid size | 265972 bp | Coordinate of oriT [Strand] | 243989..244029 [+] |
| Host baterium | Rhizobium beringeri strain TP6 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodE, nodF, nodA, nodC, nodI, nodJ, nifT, fixX, fixC, fixB, fixA, nifD, nifK, fixN, fixO, fixQ, fixP |
| Anti-CRISPR | - |