Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120363 |
| Name | oriT_pCMP234-2 |
| Organism | Klebsiella quasipneumoniae strain OSUCMP263AKPC |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP087582 (20048..20096 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pCMP234-2
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file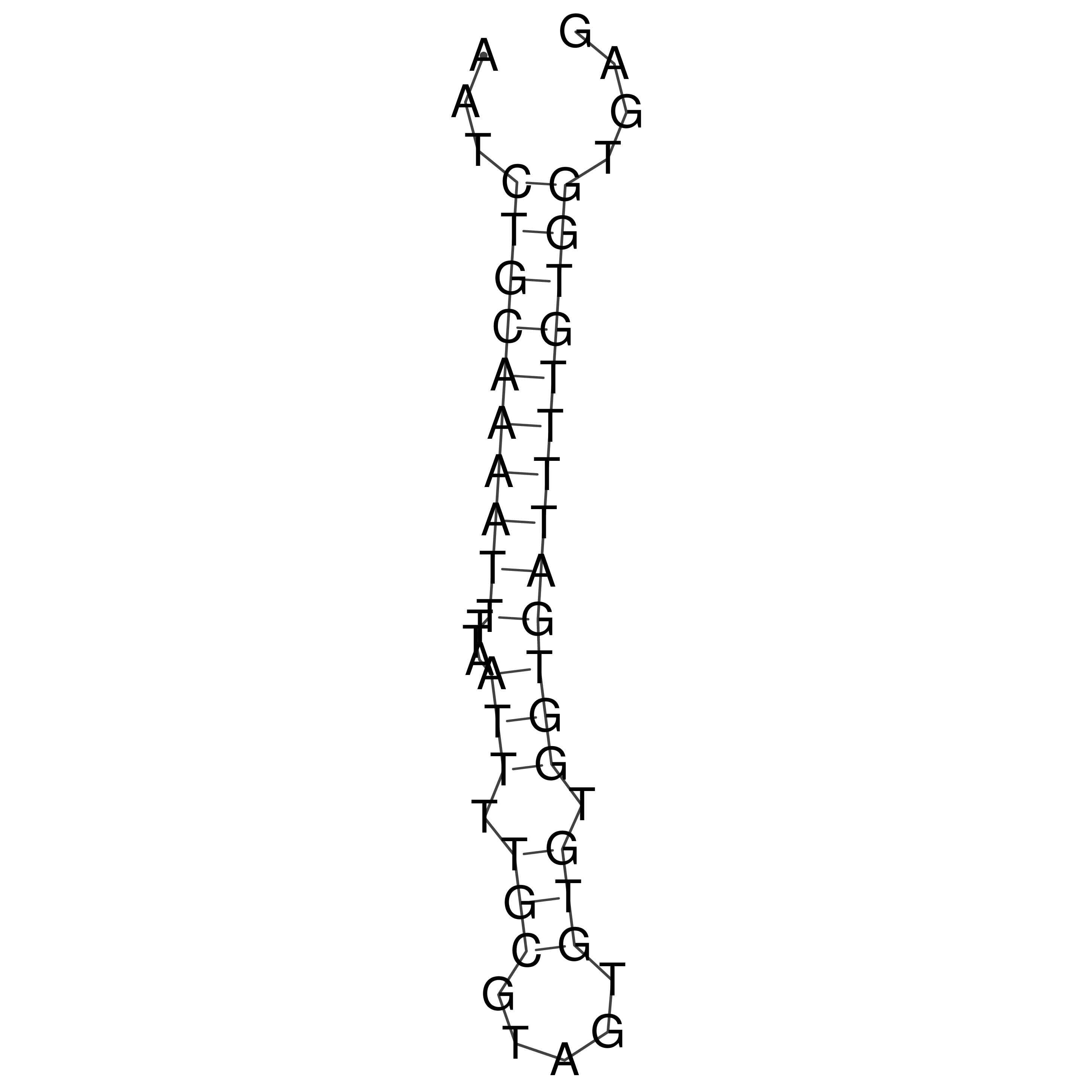
Relaxase
| ID | 12813 | GenBank | WP_032426769 |
| Name | traI_LO541_RS26115_pCMP234-2 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190629.30 Da Isoelectric Point: 6.1046
>WP_032426769.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVES
LGYRTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTSDASVVRQAPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGNHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPDSLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIQHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVES
LGYRTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTSDASVVRQAPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGNHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPDSLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIQHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7228 | GenBank | WP_032426796 |
| Name | WP_032426796_pCMP234-2 |
UniProt ID | _ |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 15036.10 Da Isoelectric Point: 4.5326
>WP_032426796.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacterales]
MGKVQAYPSDEVCEKINAIVEKRRMEGAKEKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMRTFFPDEQEEADE
MGKVQAYPSDEVCEKINAIVEKRRMEGAKEKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMRTFFPDEQEEADE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15054 | GenBank | WP_020805031 |
| Name | traD_LO541_RS26110_pCMP234-2 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85603.64 Da Isoelectric Point: 5.1733
>WP_020805031.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 22517..47805
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LO541_RS25920 (LO541_25920) | 17724..17954 | + | 231 | WP_020314756 | hypothetical protein | - |
| LO541_RS25925 (LO541_25925) | 18052..18267 | + | 216 | Protein_29 | SAM-dependent DNA methyltransferase | - |
| LO541_RS25930 (LO541_25930) | 18916..19458 | + | 543 | WP_032426797 | antirestriction protein | - |
| LO541_RS25935 (LO541_25935) | 19481..19903 | - | 423 | WP_020315406 | transglycosylase SLT domain-containing protein | - |
| LO541_RS25940 (LO541_25940) | 20409..20801 | + | 393 | WP_032426796 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LO541_RS25945 (LO541_25945) | 21039..21731 | + | 693 | WP_032426795 | conjugal transfer protein TrbJ | - |
| LO541_RS25950 (LO541_25950) | 21907..22071 | + | 165 | WP_032426810 | TraY domain-containing protein | - |
| LO541_RS25955 (LO541_25955) | 22135..22503 | + | 369 | WP_032426794 | type IV conjugative transfer system pilin TraA | - |
| LO541_RS25960 (LO541_25960) | 22517..22822 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| LO541_RS25965 (LO541_25965) | 22842..23408 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| LO541_RS25970 (LO541_25970) | 23395..24135 | + | 741 | WP_013214019 | type-F conjugative transfer system secretin TraK | traK |
| LO541_RS25975 (LO541_25975) | 24135..25559 | + | 1425 | WP_032426793 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LO541_RS25980 (LO541_25980) | 25673..26242 | + | 570 | WP_032426791 | type IV conjugative transfer system lipoprotein TraV | traV |
| LO541_RS25985 (LO541_25985) | 26397..26795 | + | 399 | WP_223294961 | hypothetical protein | - |
| LO541_RS25990 (LO541_25990) | 26824..27114 | + | 291 | WP_032426790 | hypothetical protein | - |
| LO541_RS25995 (LO541_25995) | 27138..27356 | + | 219 | WP_004171484 | hypothetical protein | - |
| LO541_RS26000 (LO541_26000) | 27357..27695 | + | 339 | WP_032426789 | hypothetical protein | - |
| LO541_RS26005 (LO541_26005) | 27740..28144 | + | 405 | WP_023320104 | hypothetical protein | - |
| LO541_RS26010 (LO541_26010) | 28169..28567 | + | 399 | WP_015065631 | hypothetical protein | - |
| LO541_RS26015 (LO541_26015) | 28639..31278 | + | 2640 | WP_032426788 | type IV secretion system protein TraC | virb4 |
| LO541_RS26020 (LO541_26020) | 31278..31667 | + | 390 | WP_032426787 | type-F conjugative transfer system protein TrbI | - |
| LO541_RS26025 (LO541_26025) | 31667..32293 | + | 627 | WP_009309871 | type-F conjugative transfer system protein TraW | traW |
| LO541_RS26030 (LO541_26030) | 32335..32724 | + | 390 | WP_004194992 | hypothetical protein | - |
| LO541_RS26035 (LO541_26035) | 32721..33710 | + | 990 | WP_009309872 | conjugal transfer pilus assembly protein TraU | traU |
| LO541_RS26040 (LO541_26040) | 33723..34361 | + | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LO541_RS26045 (LO541_26045) | 34420..36375 | + | 1956 | WP_032426778 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LO541_RS26050 (LO541_26050) | 36407..36661 | + | 255 | WP_004195500 | conjugal transfer protein TrbE | - |
| LO541_RS26055 (LO541_26055) | 36639..36887 | + | 249 | WP_004152675 | hypothetical protein | - |
| LO541_RS26060 (LO541_26060) | 36900..37226 | + | 327 | WP_004144402 | hypothetical protein | - |
| LO541_RS26065 (LO541_26065) | 37247..37999 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LO541_RS26070 (LO541_26070) | 38010..38249 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LO541_RS26075 (LO541_26075) | 38221..38778 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LO541_RS26080 (LO541_26080) | 38824..39267 | + | 444 | WP_023287131 | F-type conjugal transfer protein TrbF | - |
| LO541_RS26085 (LO541_26085) | 39245..40624 | + | 1380 | WP_072159474 | conjugal transfer pilus assembly protein TraH | traH |
| LO541_RS26090 (LO541_26090) | 40624..43446 | + | 2823 | WP_032426775 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LO541_RS26095 (LO541_26095) | 43469..44071 | + | 603 | WP_013214035 | conjugal transfer protein TraS | - |
| LO541_RS26100 (LO541_26100) | 44081..44485 | + | 405 | Protein_64 | complement resistance protein TraT | - |
| LO541_RS26105 (LO541_26105) | 44678..45367 | + | 690 | WP_023316411 | hypothetical protein | - |
| LO541_RS26110 (LO541_26110) | 45496..47805 | + | 2310 | WP_020805031 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 20792 | GenBank | NZ_CP087582 |
| Plasmid name | pCMP234-2 | Incompatibility group | - |
| Plasmid size | 104872 bp | Coordinate of oriT [Strand] | 20048..20096 [-] |
| Host baterium | Klebsiella quasipneumoniae strain OSUCMP263AKPC |
Cargo genes
| Drug resistance gene | blaKPC-2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |