Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120355 |
| Name | oriT_pSkuCIP108026b |
| Organism | Sinorhizobium kummerowiae strain CIP 108026 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP140300 (726314..726342 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSkuCIP108026b
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file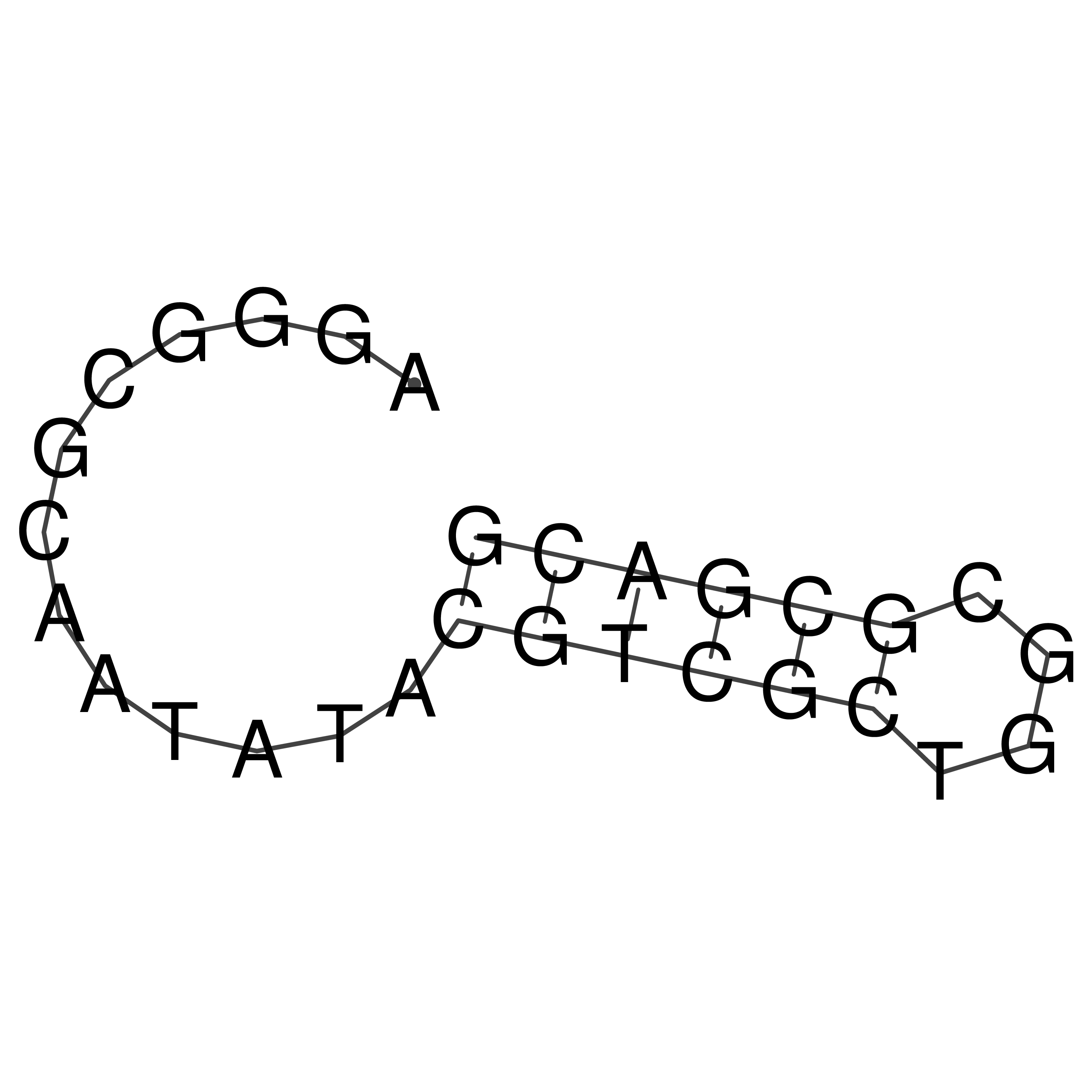
Relaxase
| ID | 12807 | GenBank | WP_284718328 |
| Name | traA_VPK21_RS10295_pSkuCIP108026b |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169883.60 Da Isoelectric Point: 9.4589
>WP_284718328.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium kummerowiae]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEALALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAFETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVAVTGEDGEPLRVITPDRPTGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGKAKQPAVFTTREMLRIE
YDMARSAEVLSKRKGFGVSNARVAAAVRSIETADTEKPFRLDPEQVDAVHHVTRDNAIAAVVGLAGAGKS
TLLAAARLAWEGDGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRETLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAISERIGFAELAGVRRQRDAWARDASRLFARGK
IEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFAALVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGVEEEVLRRWAGIAEKRELVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPLSGKHLIPPTTTFSRSVAEDARLAQLSSERWKERE
AILHPVLAKIYRDPDGALSALNALASDAAIEPRKLADDLGKAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMSLSVPALSKTAMARLVEIEAVRKQGGDDAFRTAFAF
AVEDRLLVQEVKGVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTPVDEEARTRALSAAHYRHHRVAIAETAKRIWRD
PADAIGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAAGRERRDALQALPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAADDALRRLAAQIKNKDSSLAVVAGALAPAVAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQTVRVQSSEKIVSERRQRAINQSRGIRM
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEALALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAFETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVAVTGEDGEPLRVITPDRPTGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGKAKQPAVFTTREMLRIE
YDMARSAEVLSKRKGFGVSNARVAAAVRSIETADTEKPFRLDPEQVDAVHHVTRDNAIAAVVGLAGAGKS
TLLAAARLAWEGDGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRETLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAISERIGFAELAGVRRQRDAWARDASRLFARGK
IEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFAALVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGVEEEVLRRWAGIAEKRELVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPLSGKHLIPPTTTFSRSVAEDARLAQLSSERWKERE
AILHPVLAKIYRDPDGALSALNALASDAAIEPRKLADDLGKAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMSLSVPALSKTAMARLVEIEAVRKQGGDDAFRTAFAF
AVEDRLLVQEVKGVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTPVDEEARTRALSAAHYRHHRVAIAETAKRIWRD
PADAIGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAAGRERRDALQALPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAADDALRRLAAQIKNKDSSLAVVAGALAPAVAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQTVRVQSSEKIVSERRQRAINQSRGIRM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 15048 | GenBank | WP_033048650 |
| Name | tcpA_VPK21_RS14060_pSkuCIP108026b |
UniProt ID | _ |
| Length | 947 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 947 a.a. Molecular weight: 101739.54 Da Isoelectric Point: 4.5726
>WP_033048650.1 MULTISPECIES: DNA translocase FtsK [Sinorhizobium]
MRIPRTNFSPAALYDANHADDYEDPHPAAQGAAHVPAAPEAAAHDHLFEAPEAPRRAPGHRDEETGYAGR
AARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPGWESHFFLSPNVRFTRTPERELMKRHPPAPEEN
RAEADEAAAEASDPETVMDVVPAGPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSQAPEASVTKPAI
TESVAVAEEEPATPETPSALPVTDEVPVVPHALPVANLAPESAAVEEDVPHQTDARLAYLSDFAFFEFMP
LEVAAVPPTVTEPVKDAARIPAPIAAVPKVSPPKIVAAMPVEIRPQPPTAISSLFRVVECRRPEPATAEP
VTAAPQDIAAEPAAAEAAALPAPQFVETPAPALAEPAIVPASEPAPEAPVTRAAITMPAVIQRSSPSLPP
IGALEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYE
FEPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLA
LCLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGI
PHLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGAPILATVQTGFEKGTG
EPLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIK
ANFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRP
EYLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLV
ERMEKDGLVGPANHVGKREIIYGNRDNPPKPESDDLD
MRIPRTNFSPAALYDANHADDYEDPHPAAQGAAHVPAAPEAAAHDHLFEAPEAPRRAPGHRDEETGYAGR
AARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPGWESHFFLSPNVRFTRTPERELMKRHPPAPEEN
RAEADEAAAEASDPETVMDVVPAGPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSQAPEASVTKPAI
TESVAVAEEEPATPETPSALPVTDEVPVVPHALPVANLAPESAAVEEDVPHQTDARLAYLSDFAFFEFMP
LEVAAVPPTVTEPVKDAARIPAPIAAVPKVSPPKIVAAMPVEIRPQPPTAISSLFRVVECRRPEPATAEP
VTAAPQDIAAEPAAAEAAALPAPQFVETPAPALAEPAIVPASEPAPEAPVTRAAITMPAVIQRSSPSLPP
IGALEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYE
FEPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLA
LCLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGI
PHLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGAPILATVQTGFEKGTG
EPLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIK
ANFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRP
EYLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLV
ERMEKDGLVGPANHVGKREIIYGNRDNPPKPESDDLD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 20784 | GenBank | NZ_CP140300 |
| Plasmid name | pSkuCIP108026b | Incompatibility group | - |
| Plasmid size | 1657699 bp | Coordinate of oriT [Strand] | 726314..726342 [-] |
| Host baterium | Sinorhizobium kummerowiae strain CIP 108026 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | actP |
| Degradation gene | linJ |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |