Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120262 |
| Name | oriT_LMB1|pA |
| Organism | Sinorhizobium meliloti strain LMB1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP141213 (1636796..1636824 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_LMB1|pA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file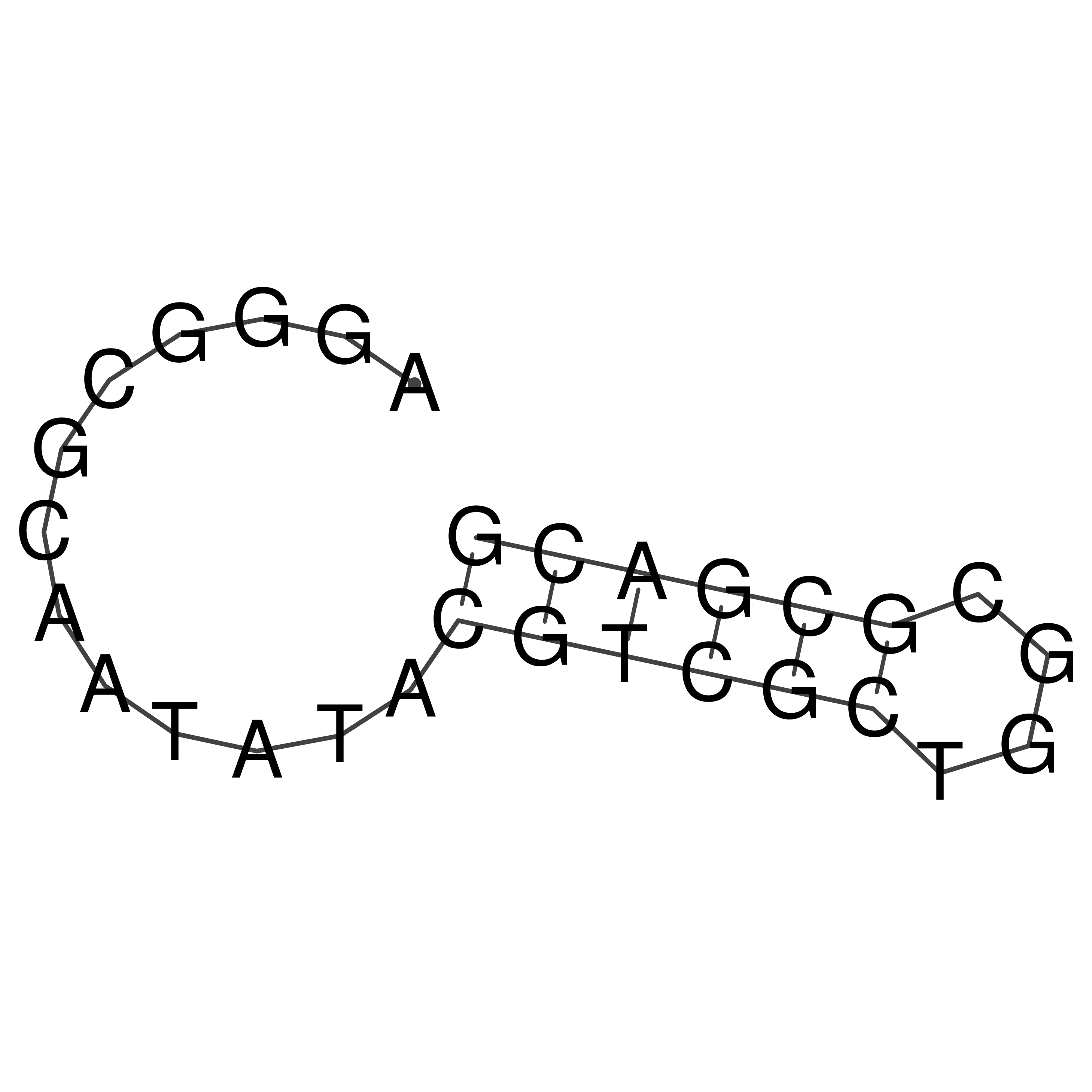
Relaxase
| ID | 12741 | GenBank | WP_324763880 |
| Name | traA_SO078_RS23920_LMB1|pA |
UniProt ID | _ |
| Length | 1541 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1541 a.a. Molecular weight: 171180.16 Da Isoelectric Point: 7.6756
>WP_324763880.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGSRELVHEELALPDDIPAWLKTAING
QLVGKAIEALWNAVDAFETRVDAQLARELIVALPEELTQAENISLVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMIPLRPLTEEGFGPKKVPVLGEHGEPLRVVTPERPNGKIVYKLWAGDKETMKAWKIAWAETANR
HLVLAGHEIRLDGRSYDEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLTEPALLFKQ
LGNERSTFDEKDIARALHRYVDDPVDFTNIRARLMASDELVMLRPQQLDAETGKASEPALFTTREILRLE
YAMAESARVLSKRRGFGVSNAQVAAAVRSIETADTEKPFKLDVEQIDAVRHVTGDNAIAAVVGLAGAGKS
TLLAAARLAWESGNRRVFGAALAGKAAEGIEDSSGIRSRTLASWELAWASGRDLLQRDDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIQAGAAFRAISERVGFAELAGVRRQRDAWARDASRLFARGK
IEEGLDTYARHGRIVAAETRDEIVDRIVADWADAREQALENSVSGGNEVRLRGDELLVLAHTNDDVRKLN
ESLRSVMVEKGALTGARAFQTARGLREFAAGDRIIFLENARFVEPRARHLGPQYVKNGMLGTVVSTGDRR
GDALLSVRLDNGRDIGISKDSYRNVDHGYATTIHKSQGATVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAATEDFEARPEWRRKPRVDHAAGVTGELVEEGIAKLRPTDEDADESPYADVRTDDGTVHRLWGVSLPKA
LEAAGVAEGDTVTLRKDGVEKVKVRVPVVDKEMGQKRFEEREVDRNVWTVRQIETADARRERIERESHRP
ELFSELVERLGRSGAKTTTLDFESEAGYWPHAKDFARRRGIDTVAEVAAEIEEGVSRRLAWLIEKREQVA
KLWERASVALGFAIERERRVSYSEERKESLSAPIPLVGKYLIPPTTKFFRSVEEDARLAQPSSERWKERE
AILRPVLAKIYRDPDGALSTLNTLASDAAIEPRKLADDLGKAPDRLGRLRGSDHMVDGRAARDERGVATV
SLHELLPLARAHATEFRRNVARFREREQTRRAHMSLSVPALSKQAMERLAEIEVVRKQGGYDAWKTAFAF
AAEDRSLVQEVKAVSDALTARFGWSAFTPKADAIVERNIDERMPDDLTDERRGKLTRLFEVVRRFAEEQD
LAERKDRSKIVAGASVEFGKEDKAVLPMLAAVTEFRTPVDEEAGPRALSAPHYRHNRAALAEIAKRIWLD
PAGAVDRIEALIVQGFAAERIVAAVANDPTAYGALRGSERLIDRMLASGRERKEALRTVSEASVFLRSLG
AAYVRAVDSERQAITEERRRMAVAIPGLSQAAEDALLRLTAEMKKKDGKLDVAAGALAPAVAREFAAVSR
ALDERFGRNAILRGETDVINRVPPPHRRAFEAMQDRLKILQETVRMQASQQIISERQQRIIDRARGEACG
N
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGSRELVHEELALPDDIPAWLKTAING
QLVGKAIEALWNAVDAFETRVDAQLARELIVALPEELTQAENISLVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMIPLRPLTEEGFGPKKVPVLGEHGEPLRVVTPERPNGKIVYKLWAGDKETMKAWKIAWAETANR
HLVLAGHEIRLDGRSYDEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLTEPALLFKQ
LGNERSTFDEKDIARALHRYVDDPVDFTNIRARLMASDELVMLRPQQLDAETGKASEPALFTTREILRLE
YAMAESARVLSKRRGFGVSNAQVAAAVRSIETADTEKPFKLDVEQIDAVRHVTGDNAIAAVVGLAGAGKS
TLLAAARLAWESGNRRVFGAALAGKAAEGIEDSSGIRSRTLASWELAWASGRDLLQRDDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIQAGAAFRAISERVGFAELAGVRRQRDAWARDASRLFARGK
IEEGLDTYARHGRIVAAETRDEIVDRIVADWADAREQALENSVSGGNEVRLRGDELLVLAHTNDDVRKLN
ESLRSVMVEKGALTGARAFQTARGLREFAAGDRIIFLENARFVEPRARHLGPQYVKNGMLGTVVSTGDRR
GDALLSVRLDNGRDIGISKDSYRNVDHGYATTIHKSQGATVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAATEDFEARPEWRRKPRVDHAAGVTGELVEEGIAKLRPTDEDADESPYADVRTDDGTVHRLWGVSLPKA
LEAAGVAEGDTVTLRKDGVEKVKVRVPVVDKEMGQKRFEEREVDRNVWTVRQIETADARRERIERESHRP
ELFSELVERLGRSGAKTTTLDFESEAGYWPHAKDFARRRGIDTVAEVAAEIEEGVSRRLAWLIEKREQVA
KLWERASVALGFAIERERRVSYSEERKESLSAPIPLVGKYLIPPTTKFFRSVEEDARLAQPSSERWKERE
AILRPVLAKIYRDPDGALSTLNTLASDAAIEPRKLADDLGKAPDRLGRLRGSDHMVDGRAARDERGVATV
SLHELLPLARAHATEFRRNVARFREREQTRRAHMSLSVPALSKQAMERLAEIEVVRKQGGYDAWKTAFAF
AAEDRSLVQEVKAVSDALTARFGWSAFTPKADAIVERNIDERMPDDLTDERRGKLTRLFEVVRRFAEEQD
LAERKDRSKIVAGASVEFGKEDKAVLPMLAAVTEFRTPVDEEAGPRALSAPHYRHNRAALAEIAKRIWLD
PAGAVDRIEALIVQGFAAERIVAAVANDPTAYGALRGSERLIDRMLASGRERKEALRTVSEASVFLRSLG
AAYVRAVDSERQAITEERRRMAVAIPGLSQAAEDALLRLTAEMKKKDGKLDVAAGALAPAVAREFAAVSR
ALDERFGRNAILRGETDVINRVPPPHRRAFEAMQDRLKILQETVRMQASQQIISERQQRIIDRARGEACG
N
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14958 | GenBank | WP_324764498 |
| Name | tcpA_SO078_RS19785_LMB1|pA |
UniProt ID | _ |
| Length | 946 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 946 a.a. Molecular weight: 101511.00 Da Isoelectric Point: 4.5462
>WP_324764498.1 DNA translocase FtsK [Sinorhizobium meliloti]
MRIPRTNFSPAALYDANHAVDFEEPHPVAQGSADASAPEAAAHDHLFEAPEAPRRAPGHRDQETGYAGRA
ARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPSWESHFFLSPNVRFTRTPERELMKRHPPAPEESR
AEADEAAAEASDPETVMDAVPAEPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSPAPEASTTEPAIT
ESAAVTEEEPTTPETSSALPVADEVPVVPHALPVANLAPRSAGVEEDVPQQAEAHLAYLSDFAFFEFMPL
EAAAVPTAAAEPVKEAAHLRAPVAAAPKVSPPKIAAATPVEIRPQPPTAISSLFRVVECRRPEPAAAEPA
SAAPQDLAAEPTVVETAAPVIAEAPASAITEPLVPGVVPANEPPPEVPVTRAAITMPAVIQRSSPSLPPI
GAIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEF
EPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLAL
CLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGIP
HLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGEPILATVQTGFEKGTGE
PLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIKA
NFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRPE
YLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVE
RMEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
MRIPRTNFSPAALYDANHAVDFEEPHPVAQGSADASAPEAAAHDHLFEAPEAPRRAPGHRDQETGYAGRA
ARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPSWESHFFLSPNVRFTRTPERELMKRHPPAPEESR
AEADEAAAEASDPETVMDAVPAEPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSPAPEASTTEPAIT
ESAAVTEEEPTTPETSSALPVADEVPVVPHALPVANLAPRSAGVEEDVPQQAEAHLAYLSDFAFFEFMPL
EAAAVPTAAAEPVKEAAHLRAPVAAAPKVSPPKIAAATPVEIRPQPPTAISSLFRVVECRRPEPAAAEPA
SAAPQDLAAEPTVVETAAPVIAEAPASAITEPLVPGVVPANEPPPEVPVTRAAITMPAVIQRSSPSLPPI
GAIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEF
EPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLAL
CLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGIP
HLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGEPILATVQTGFEKGTGE
PLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIKA
NFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRPE
YLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVE
RMEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14959 | GenBank | WP_324763882 |
| Name | traG_SO078_RS23935_LMB1|pA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70444.27 Da Isoelectric Point: 9.5745
>WP_324763882.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MTVRAKPHPSLLLVLVPIAVTAVAVYVVGWRWPTLAGGLSGKAEYWFLRASPVPALLFGPLAGLLSVWAL
PLHRRRPVAMASLVCFLAVAGFYALREFGRLAPSVESRAVTWDRALSYLDMVAVFSAVAGFMAAAVAARI
STVVPEPVKRARRGTFGDADWLPMSAAGKLFPPEGEIVVGERYRVDKDIVHGLPFDPNDRSTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRSRVLGREVMVLDPT
NPTMGFNVLDGIEASKRQEEDIVGLAHMLLSESARFESSSGAYFQNQAHNLLTGLLAHVMFSPDFEGRRN
LRSLRQIVSEPEPSVLAMLRDVQEHSHSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GSAFKSSDIVTAKKDVFLNIPATILRSYPGIGRVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSLGQLERHFGRDGATSWIDGCALASYAAIKAFDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNGASRKSESVNYQRRPLIMPHEITHSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMKEAAK
ANRFVKPIP
MTVRAKPHPSLLLVLVPIAVTAVAVYVVGWRWPTLAGGLSGKAEYWFLRASPVPALLFGPLAGLLSVWAL
PLHRRRPVAMASLVCFLAVAGFYALREFGRLAPSVESRAVTWDRALSYLDMVAVFSAVAGFMAAAVAARI
STVVPEPVKRARRGTFGDADWLPMSAAGKLFPPEGEIVVGERYRVDKDIVHGLPFDPNDRSTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRSRVLGREVMVLDPT
NPTMGFNVLDGIEASKRQEEDIVGLAHMLLSESARFESSSGAYFQNQAHNLLTGLLAHVMFSPDFEGRRN
LRSLRQIVSEPEPSVLAMLRDVQEHSHSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GSAFKSSDIVTAKKDVFLNIPATILRSYPGIGRVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSLGQLERHFGRDGATSWIDGCALASYAAIKAFDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNGASRKSESVNYQRRPLIMPHEITHSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMKEAAK
ANRFVKPIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1642606..1652177
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SO078_RS23940 (SO078_23940) | 1640118..1640333 | + | 216 | Protein_1494 | WGR domain-containing protein | - |
| SO078_RS23945 (SO078_23945) | 1640490..1640657 | - | 168 | WP_324763883 | hypothetical protein | - |
| SO078_RS23950 (SO078_23950) | 1641009..1641332 | - | 324 | WP_324763884 | DUF736 family protein | - |
| SO078_RS23955 (SO078_23955) | 1642057..1642479 | + | 423 | WP_324764648 | hypothetical protein | - |
| SO078_RS23960 (SO078_23960) | 1642606..1643595 | - | 990 | WP_324763885 | P-type DNA transfer ATPase VirB11 | virB11 |
| SO078_RS23965 (SO078_23965) | 1643606..1644775 | - | 1170 | WP_324763886 | type IV secretion system protein VirB10 | virB10 |
| SO078_RS23970 (SO078_23970) | 1644785..1645639 | - | 855 | WP_324763887 | P-type conjugative transfer protein VirB9 | virB9 |
| SO078_RS23975 (SO078_23975) | 1645639..1646310 | - | 672 | WP_324763888 | virB8 family protein | virB8 |
| SO078_RS23980 (SO078_23980) | 1646315..1646590 | - | 276 | WP_324763889 | hypothetical protein | - |
| SO078_RS23985 (SO078_23985) | 1646627..1647562 | - | 936 | WP_324763890 | type IV secretion system protein | virB6 |
| SO078_RS23990 (SO078_23990) | 1647583..1647798 | - | 216 | WP_324763891 | EexN family lipoprotein | - |
| SO078_RS23995 (SO078_23995) | 1647795..1648493 | - | 699 | WP_324764634 | P-type DNA transfer protein VirB5 | virB5 |
| SO078_RS24000 (SO078_24000) | 1648493..1650874 | - | 2382 | WP_324763892 | VirB4 family type IV secretion system protein | virb4 |
| SO078_RS24005 (SO078_24005) | 1650867..1651208 | - | 342 | WP_324763893 | type IV secretion system protein VirB3 | virB3 |
| SO078_RS24010 (SO078_24010) | 1651213..1651512 | - | 300 | WP_324763894 | TrbC/VirB2 family protein | virB2 |
| SO078_RS24015 (SO078_24015) | 1651509..1652177 | - | 669 | WP_324763895 | type IV secretion system protein VirB1 | virB1 |
| SO078_RS24020 (SO078_24020) | 1652181..1652699 | - | 519 | WP_324763896 | hypothetical protein | - |
| SO078_RS24025 (SO078_24025) | 1652796..1653164 | + | 369 | WP_324763897 | MarR family transcriptional regulator | - |
| SO078_RS24030 (SO078_24030) | 1654420..1656723 | - | 2304 | WP_324763898 | AGE family epimerase/isomerase | - |
Host bacterium
| ID | 20691 | GenBank | NZ_CP141213 |
| Plasmid name | LMB1|pA | Incompatibility group | - |
| Plasmid size | 1820429 bp | Coordinate of oriT [Strand] | 1636796..1636824 [-] |
| Host baterium | Sinorhizobium meliloti strain LMB1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | actP |
| Degradation gene | RPYSC3_16500 |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |