Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120253 |
| Name | oriT_K377|unnamed1 |
| Organism | Agrobacterium vitis strain K377 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JACXXJ020000001 (113811..113851 [-], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_K377|unnamed1
TCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
TCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file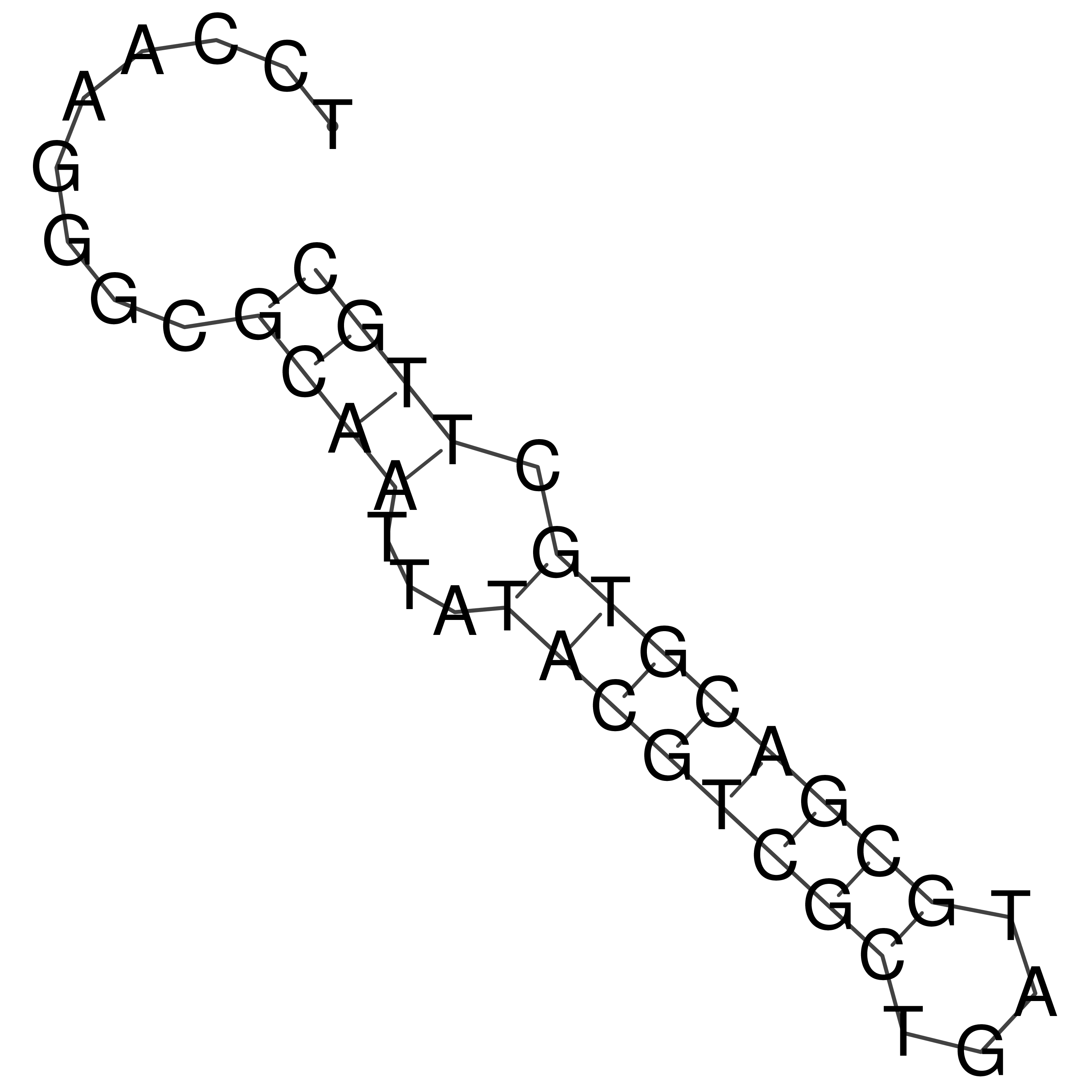
Relaxase
| ID | 12739 | GenBank | WP_194415504 |
| Name | traA_IEI95_RS00520_K377|unnamed1 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 124074.89 Da Isoelectric Point: 9.8011
>WP_194415504.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium vitis]
MAIAHFSASIVSRGGGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLVHEEFMLPADAPKWVRAMLADR
SVAGASEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIVLVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEEGFGPKKVAVIGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLKIDGRSYEKQGIELEPTIHLGVGAKAIERKAQEQGVRPELERLELNEERRSENARRILRRPEIVLDL
ITREKSVFDERDVAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIDFSTGEKVPARYSTRAMIRLEA
TMMRQALWLSNRDGHAVSEAALDATFRRHERLSEEQKTAIERIAGPARIAAVVGRAGAGKTTMMKAAREV
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDKTVFVMDEAGMVASKQMAGFVD
TVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNIMAREKLVERGIVGEGHVFKTADG
IRQFDVGDQIVFLKNETSLGVKNGMIAHVIEAAPNRIVAVVGEGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDCVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFSFNGGLSKVLSRRNAKETTLDYERGRLY
REALRFAENRGLHIMQVARTLLRDRLDWTLRQKTKLSDLVHRLRALGERFGLDQSPKTQTMKEAAPMVAG
IKTFSGSVADTLSDKLGADPALKQQWEEVSARFRYVFADPETAFRAMNFDAVLADKDSARHALQKLEAEP
ASIGALKGKTGILASKTEREARRIAELNVPALKRDLVQYLRMRETATQRMATEEQALRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTERFGERSLLTNAAREPFGKLFDKFSDG
MQPEQKERLKEAWPIMRTAQQIAAHERTVQTLRQSEGLRLNQRQTPVLKQ
MAIAHFSASIVSRGGGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLVHEEFMLPADAPKWVRAMLADR
SVAGASEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIVLVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEEGFGPKKVAVIGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLKIDGRSYEKQGIELEPTIHLGVGAKAIERKAQEQGVRPELERLELNEERRSENARRILRRPEIVLDL
ITREKSVFDERDVAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIDFSTGEKVPARYSTRAMIRLEA
TMMRQALWLSNRDGHAVSEAALDATFRRHERLSEEQKTAIERIAGPARIAAVVGRAGAGKTTMMKAAREV
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDKTVFVMDEAGMVASKQMAGFVD
TVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNIMAREKLVERGIVGEGHVFKTADG
IRQFDVGDQIVFLKNETSLGVKNGMIAHVIEAAPNRIVAVVGEGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDCVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFSFNGGLSKVLSRRNAKETTLDYERGRLY
REALRFAENRGLHIMQVARTLLRDRLDWTLRQKTKLSDLVHRLRALGERFGLDQSPKTQTMKEAAPMVAG
IKTFSGSVADTLSDKLGADPALKQQWEEVSARFRYVFADPETAFRAMNFDAVLADKDSARHALQKLEAEP
ASIGALKGKTGILASKTEREARRIAELNVPALKRDLVQYLRMRETATQRMATEEQALRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTERFGERSLLTNAAREPFGKLFDKFSDG
MQPEQKERLKEAWPIMRTAQQIAAHERTVQTLRQSEGLRLNQRQTPVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14953 | GenBank | WP_194415643 |
| Name | t4cp2_IEI95_RS00065_K377|unnamed1 |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 91983.72 Da Isoelectric Point: 5.5934
>WP_194415643.1 conjugal transfer protein TrbE [Agrobacterium vitis]
MVALKPFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDVERNEVSRQINMILSRLG
SGWMIQVEALRVPTIDYAEEDACHFRDPVTRAIDTERRSHFERESGHFESRHALILTWRAPEPRRSGMAR
YVYSDTASRSATYADTALESFRTSIREVEQYLANVVSIRRMTTRETEERGGFRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPTVENRFLGVVAIDGLAAESWPGILNSLDLMPLTYRWSSRFVF
LDEQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAQAASQLVAYGYYTPVIVL
FDDQQPRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPIAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLSLIAAQFRRYAGAQIFA
FDKGRSMLPLTLAIGGDHYEIGGDGAGEGDGASPRLAFCPLAELSTDSDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGAFQCFEVEEL
MNLGERNLVPVLLYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPVALSFVGASGKEDLKRIRALHSEYGADWPKHWLQQRGIAHVETLFPAA
MVALKPFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDVERNEVSRQINMILSRLG
SGWMIQVEALRVPTIDYAEEDACHFRDPVTRAIDTERRSHFERESGHFESRHALILTWRAPEPRRSGMAR
YVYSDTASRSATYADTALESFRTSIREVEQYLANVVSIRRMTTRETEERGGFRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPTVENRFLGVVAIDGLAAESWPGILNSLDLMPLTYRWSSRFVF
LDEQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAQAASQLVAYGYYTPVIVL
FDDQQPRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPIAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLSLIAAQFRRYAGAQIFA
FDKGRSMLPLTLAIGGDHYEIGGDGAGEGDGASPRLAFCPLAELSTDSDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGAFQCFEVEEL
MNLGERNLVPVLLYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPVALSFVGASGKEDLKRIRALHSEYGADWPKHWLQQRGIAHVETLFPAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14954 | GenBank | WP_194415508 |
| Name | traG_IEI95_RS00535_K377|unnamed1 |
UniProt ID | _ |
| Length | 658 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 658 a.a. Molecular weight: 71215.59 Da Isoelectric Point: 9.3596
>WP_194415508.1 Ti-type conjugative transfer system protein TraG [Agrobacterium vitis]
MTMNRLLLLILPAIIMLAAMFATSGMEQRLAAFGTSPQAKLMLGRAGLALPYIAAVAIGIIGLFAANGWA
NIKAAGLSVLAGNGVVITIATLREVIRLNSIASRVPAEQSVLAYADPVTMIGVGTAFISGMFALRVAIKG
NAAFATTAPKRIGGKRALHGEADWMKIQEAAKLFPEPGGIVIGERYRVDRDSVAAMPFRADDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVCEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTEV
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGNVKGRTLFLLDEVARLGY
LRILETARDAGRKYGIMLTTIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSTGMKGSSRSRSKQLSRRPLILPHEVLRMRSDEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
ENRFHRTGTGTDTHTEHAPPWQKEGTRP
MTMNRLLLLILPAIIMLAAMFATSGMEQRLAAFGTSPQAKLMLGRAGLALPYIAAVAIGIIGLFAANGWA
NIKAAGLSVLAGNGVVITIATLREVIRLNSIASRVPAEQSVLAYADPVTMIGVGTAFISGMFALRVAIKG
NAAFATTAPKRIGGKRALHGEADWMKIQEAAKLFPEPGGIVIGERYRVDRDSVAAMPFRADDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVCEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTEV
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGNVKGRTLFLLDEVARLGY
LRILETARDAGRKYGIMLTTIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSTGMKGSSRSRSKQLSRRPLILPHEVLRMRSDEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
ENRFHRTGTGTDTHTEHAPPWQKEGTRP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 11606..21278
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IEI95_RS00030 (IEI95_000030) | 6849..8168 | - | 1320 | WP_194415587 | plasmid replication protein RepC | - |
| IEI95_RS00035 (IEI95_000035) | 8395..9396 | - | 1002 | WP_194415589 | plasmid partitioning protein RepB | - |
| IEI95_RS00040 (IEI95_000040) | 9393..10604 | - | 1212 | WP_194415590 | plasmid partitioning protein RepA | - |
| IEI95_RS00045 (IEI95_000045) | 10974..11609 | + | 636 | WP_194415592 | acyl-homoserine-lactone synthase | - |
| IEI95_RS00050 (IEI95_000050) | 11606..12577 | + | 972 | WP_194415594 | P-type conjugative transfer ATPase TrbB | virB11 |
| IEI95_RS00055 (IEI95_000055) | 12567..12971 | + | 405 | WP_194415596 | conjugal transfer pilin TrbC | virB2 |
| IEI95_RS00060 (IEI95_000060) | 12964..13263 | + | 300 | WP_045025167 | conjugal transfer protein TrbD | virB3 |
| IEI95_RS00065 (IEI95_000065) | 13274..15742 | + | 2469 | WP_194415643 | conjugal transfer protein TrbE | virb4 |
| IEI95_RS00070 (IEI95_000070) | 15714..16523 | + | 810 | WP_194415599 | P-type conjugative transfer protein TrbJ | virB5 |
| IEI95_RS00075 (IEI95_000075) | 16520..16738 | + | 219 | WP_194415601 | entry exclusion protein TrbK | - |
| IEI95_RS00080 (IEI95_000080) | 16732..17928 | + | 1197 | WP_194415603 | P-type conjugative transfer protein TrbL | virB6 |
| IEI95_RS00085 (IEI95_000085) | 17942..18604 | + | 663 | WP_071208211 | conjugal transfer protein TrbF | virB8 |
| IEI95_RS00090 (IEI95_000090) | 18622..19476 | + | 855 | WP_071208371 | P-type conjugative transfer protein TrbG | virB9 |
| IEI95_RS00095 (IEI95_000095) | 19476..19955 | + | 480 | WP_194415605 | conjugal transfer protein TrbH | - |
| IEI95_RS00100 (IEI95_000100) | 19968..21278 | + | 1311 | WP_194415607 | IncP-type conjugal transfer protein TrbI | virB10 |
| IEI95_RS00105 (IEI95_000105) | 21332..23197 | - | 1866 | WP_194415609 | LysR family transcriptional regulator | - |
| IEI95_RS00110 (IEI95_000110) | 23368..24705 | - | 1338 | WP_234934142 | MmgE/PrpD family protein | - |
| IEI95_RS00115 (IEI95_000115) | 24851..25636 | - | 786 | WP_194415614 | amino acid ABC transporter ATP-binding protein | - |
Host bacterium
| ID | 20682 | GenBank | NZ_JACXXJ020000001 |
| Plasmid name | K377|unnamed1 | Incompatibility group | - |
| Plasmid size | 187327 bp | Coordinate of oriT [Strand] | 113811..113851 [-] |
| Host baterium | Agrobacterium vitis strain K377 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF7 |