Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120226 |
| Name | oriT_Dcc0527|unnamed4 |
| Organism | Cronobacter malonaticus strain Dcc0527 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP141996 (55333..55413 [-], 81 nt) |
| oriT length | 81 nt |
| IRs (inverted repeats) | 61..68, 73..80 (TTGGTGGT..ACCACCAA) 27..34, 37..44 (GCAAAAAC..GTTTTTGC) 8..13, 21..26 (TGATTT..AAATCA) |
| Location of nic site | 53..54 |
| Conserved sequence flanking the nic site |
TGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 81 nt
AATTACATGATTTAAAACGCAAATCAGCAAAAACTTGTTTTTGCGTAGTGTGTGGTGCTTTTGGTGGTGAGAACCACCAAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file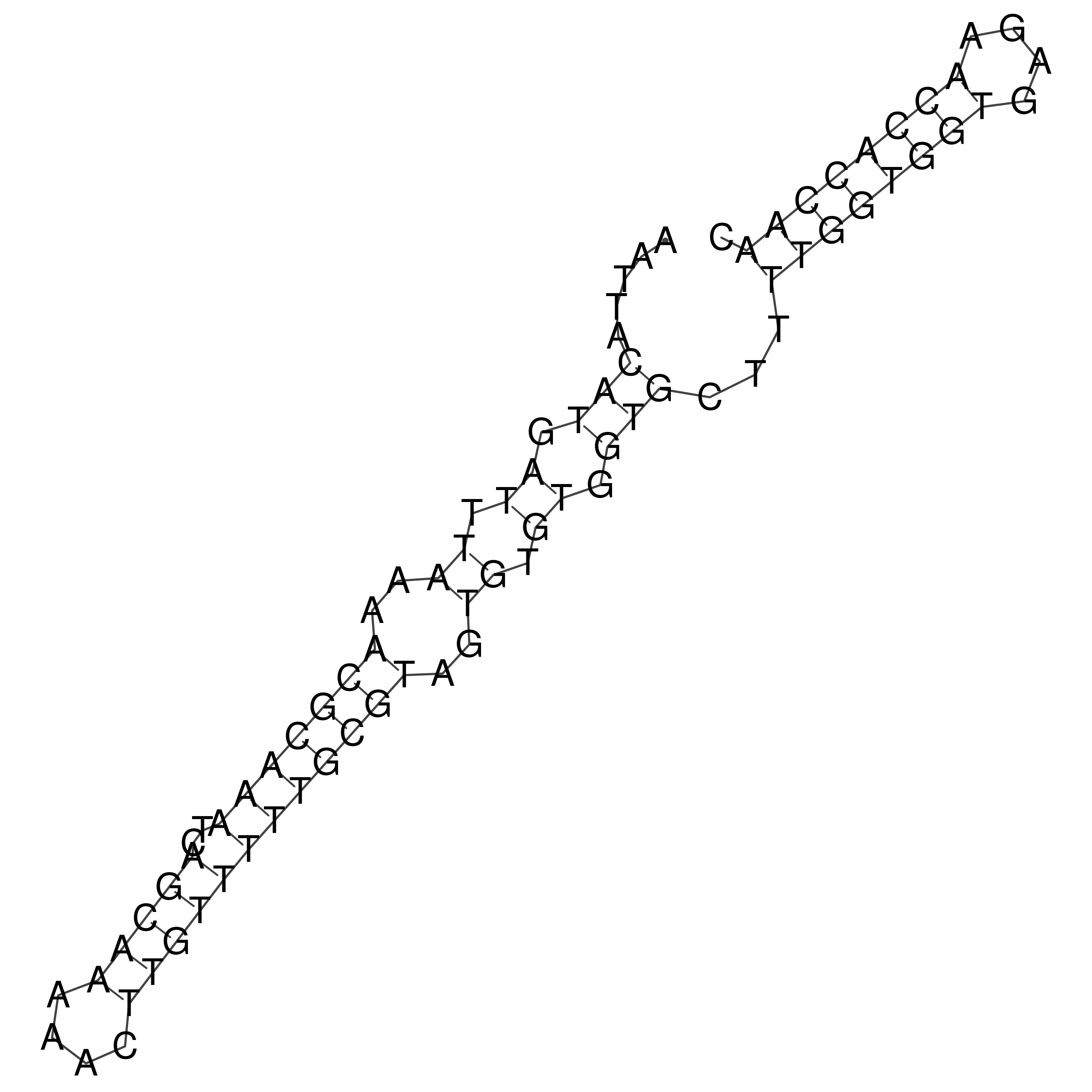
Relaxase
| ID | 12725 | GenBank | WP_324688948 |
| Name | traI_U9L39_RS21880_Dcc0527|unnamed4 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192040.64 Da Isoelectric Point: 5.9775
MLSFSVVKSAGSAGNYYTDKDNYYVLGSMGERWAGQGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQDPQLHTHVVVANVTQHNGEWKTLSSDKVGKTGFSENVLANRIAFGKIYQSELRQRVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEIRMAEWMQT
LKETGFDIRAYRDAADQRAEIRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRQEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDVMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDKATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISHQKSALNTPAQQAIHLALPVVESKNLAFSHVDLLTEAKSFAAEGTSFADLGREINA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETPDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYTLIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWVPEHSVTEFSHSQEAKLAEA
QQEAMQKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPAQEQAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTSDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVREIAGQERDRAAITEREAALPESVLREPQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7178 | GenBank | WP_021559009 |
| Name | WP_021559009_Dcc0527|unnamed4 |
UniProt ID | _ |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14493.50 Da Isoelectric Point: 5.0120
MARVILYISNDVYDKVNAIVEQRRQEGARDKDISISGTASMLLELGLRVHQAQMERKESAFNQTEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSVEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 7179 | GenBank | WP_000089265 |
| Name | WP_000089265_Dcc0527|unnamed4 |
UniProt ID | A0A1E5M428 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 8472.68 Da Isoelectric Point: 9.9218
MSRNIIRPAPGNKVLLVLDDATNHKLLGARERSGRTKTNEVLVRLRDHLSRFPDFYNLDAIKEGTGETDS
IIKDL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1E5M428 |
T4CP
| ID | 14938 | GenBank | WP_021519755 |
| Name | traC_U9L39_RS21770_Dcc0527|unnamed4 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99327.25 Da Isoelectric Point: 6.5173
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASFHGAKITTQTVDAQAFIEIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPEMSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFKGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVKASWLAKKNQARIDDVVNFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRTLKKLNVIDEGWR
LLDFKNRKVGEFIQKGYRTCRRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLFPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRREGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14939 | GenBank | WP_034169381 |
| Name | traD_U9L39_RS21875_Dcc0527|unnamed4 |
UniProt ID | _ |
| Length | 738 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 738 a.a. Molecular weight: 83846.52 Da Isoelectric Point: 5.2346
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLILWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPEVASGEGVTQAEQPQQPQQPQQPQQ
PQQPQQPQQPQQPQQPVSSVINDKKSDAGVSVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYE
AWQQENHPDIQQHMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 366..21323
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| U9L39_RS21755 (U9L39_21755) | 49..366 | + | 318 | WP_024189706 | conjugal transfer protein TrbD | - |
| U9L39_RS21760 (U9L39_21760) | 366..881 | + | 516 | WP_021519754 | type IV conjugative transfer system lipoprotein TraV | traV |
| U9L39_RS21765 (U9L39_21765) | 1016..1237 | + | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| U9L39_RS21770 (U9L39_21770) | 1397..4024 | + | 2628 | WP_021519755 | type IV secretion system protein TraC | virb4 |
| U9L39_RS21775 (U9L39_21775) | 4021..4407 | + | 387 | WP_000214096 | type-F conjugative transfer system protein TrbI | - |
| U9L39_RS21780 (U9L39_21780) | 4404..5036 | + | 633 | WP_062873969 | type-F conjugative transfer system protein TraW | traW |
| U9L39_RS21785 (U9L39_21785) | 5033..6025 | + | 993 | WP_021559016 | conjugal transfer pilus assembly protein TraU | traU |
| U9L39_RS21790 (U9L39_21790) | 6055..6360 | + | 306 | WP_122987481 | hypothetical protein | - |
| U9L39_RS21795 (U9L39_21795) | 6369..7007 | + | 639 | WP_032083946 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| U9L39_RS21800 (U9L39_21800) | 7004..7384 | + | 381 | WP_324688930 | hypothetical protein | - |
| U9L39_RS21805 (U9L39_21805) | 7620..9482 | + | 1863 | WP_226149361 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| U9L39_RS21810 (U9L39_21810) | 9509..9766 | + | 258 | WP_000864336 | conjugal transfer protein TrbE | - |
| U9L39_RS21815 (U9L39_21815) | 9759..10502 | + | 744 | WP_130570191 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| U9L39_RS21820 (U9L39_21820) | 10663..10899 | + | 237 | WP_000270010 | type-F conjugative transfer system pilin chaperone TraQ | - |
| U9L39_RS21825 (U9L39_21825) | 10886..11431 | + | 546 | WP_000059817 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| U9L39_RS21830 (U9L39_21830) | 11421..11735 | + | 315 | WP_001384255 | P-type conjugative transfer protein TrbJ | - |
| U9L39_RS21835 (U9L39_21835) | 11689..12081 | + | 393 | WP_000126338 | F-type conjugal transfer protein TrbF | - |
| U9L39_RS21840 (U9L39_21840) | 12068..12241 | + | 174 | Protein_18 | conjugal transfer protein TraH | - |
| U9L39_RS21845 (U9L39_21845) | 12268..12660 | + | 393 | WP_000660703 | F-type conjugal transfer protein TrbF | - |
| U9L39_RS21850 (U9L39_21850) | 12647..14020 | + | 1374 | WP_001137363 | conjugal transfer pilus assembly protein TraH | traH |
| U9L39_RS21855 (U9L39_21855) | 14017..16836 | + | 2820 | WP_032214174 | conjugal transfer mating-pair stabilization protein TraG | traG |
| U9L39_RS21860 (U9L39_21860) | 16855..17352 | + | 498 | WP_000605862 | entry exclusion protein | - |
| U9L39_RS21865 (U9L39_21865) | 17385..18116 | + | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| U9L39_RS21870 (U9L39_21870) | 18319..19056 | + | 738 | WP_000199911 | hypothetical protein | - |
| U9L39_RS21875 (U9L39_21875) | 19107..21323 | + | 2217 | WP_034169381 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 20655 | GenBank | NZ_CP141996 |
| Plasmid name | Dcc0527|unnamed4 | Incompatibility group | - |
| Plasmid size | 61266 bp | Coordinate of oriT [Strand] | 55333..55413 [-] |
| Host baterium | Cronobacter malonaticus strain Dcc0527 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |