Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120194 |
| Name | oriT_6S-65-1|p1 |
| Organism | Escherichia albertii strain 6S-65-1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP141902 (67772..67862 [-], 91 nt) |
| oriT length | 91 nt |
| IRs (inverted repeats) | 66..73, 78..85 (TTGGTGGT..ACCACCAA) 27..34, 37..44 (GCAAAAAC..GTTTTTGC) 8..13, 21..26 (TGATTT..AAATCA) |
| Location of nic site | 53..54 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 91 nt
AATTACATGATTTAAAACACAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCGGTGCTTTTGGTGGTGAGAACCACCAACCTGTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file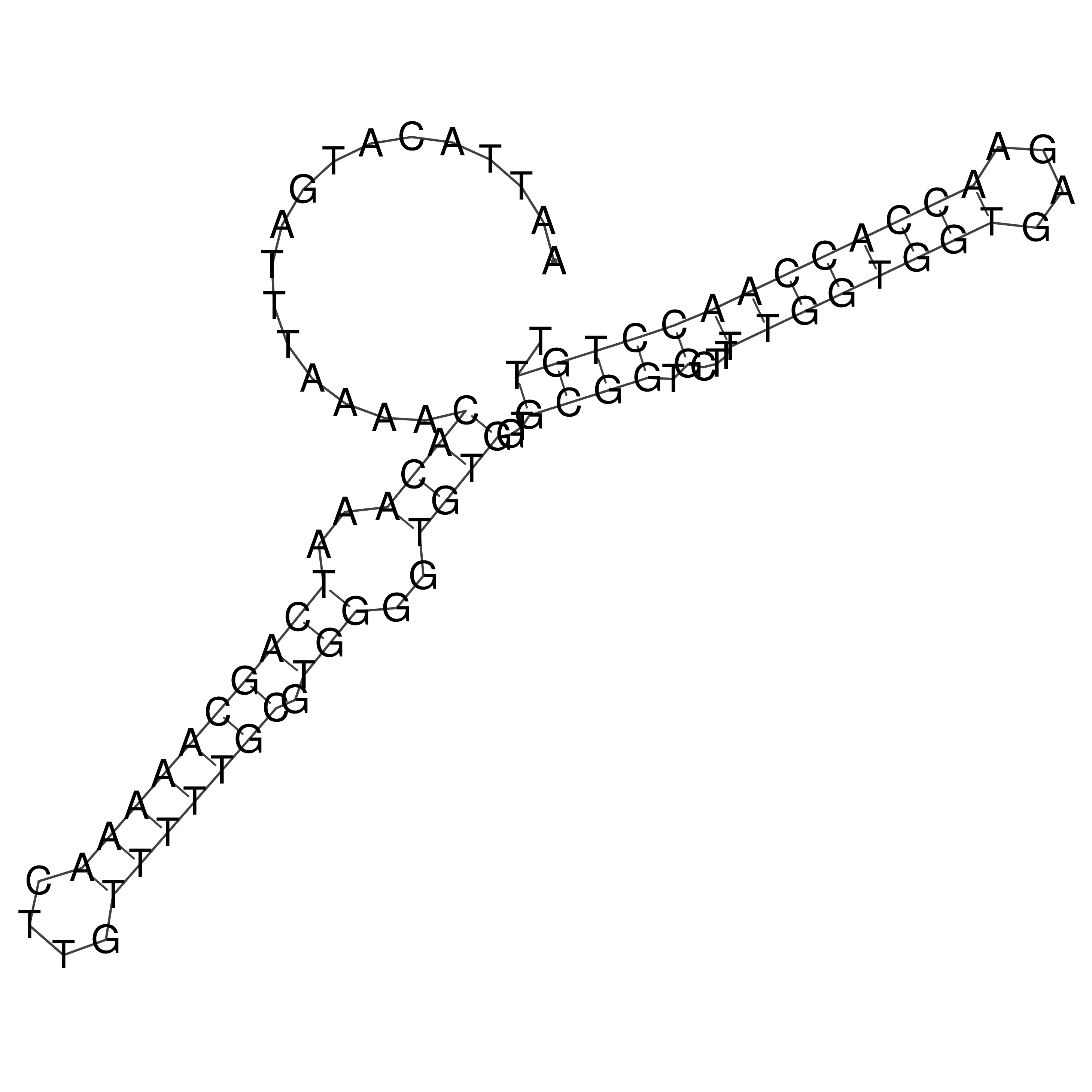
Relaxase
| ID | 12705 | GenBank | WP_324649223 |
| Name | traI_VK940_RS24750_6S-65-1|p1 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192342.96 Da Isoelectric Point: 6.0786
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEGASLKSRDVAALDTRKSKQHVDPEIRMAEWMQT
LKETGFDIRAYRDAADQRAEIRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQDGMTFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQILITDSGQRTGTGSALMSMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQSIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPATLPVADSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARNPSFTVVSEQI
KARAGETLLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSQVDLLTEAKSFAAEGTSFVDLGREIDA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDHTGRTPETREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQKTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKSDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGRTEQAVREIAWQERERAVTSEREAALPESVLREPQREREAVREVVRENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7158 | GenBank | WP_001063020 |
| Name | WP_001063020_6S-65-1|p1 |
UniProt ID | A0A6Y3HD60 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14442.54 Da Isoelectric Point: 5.3125
MAKIQVYVNDHVSEKINAIAVQRRAEGAKEKDVSYSSIASMLLELGLRVYEAQMERKESAFNQAEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGKPKFEYANMVEDIREKVSVEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A6Y3HD60 |
| ID | 7159 | GenBank | WP_024226173 |
| Name | WP_024226173_6S-65-1|p1 |
UniProt ID | _ |
| Length | 71 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 71 a.a. Molecular weight: 8131.22 Da Isoelectric Point: 9.9216
MSRGRTRAAPGNKVLLILDETTNQKLLAARDRSGRTKTNEVFIRLKDHLNRFPDFYNASLIKEEAESIDD
I
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14911 | GenBank | WP_024226180 |
| Name | traC_VK940_RS24655_6S-65-1|p1 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99211.87 Da Isoelectric Point: 5.7326
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQMVGDRIEYGLREFSWSGEQAERFNAITQAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVTDNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 67204..89122
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| VK940_RS24545 (VK940_24545) | 63200..63634 | + | 435 | WP_000845950 | conjugation system SOS inhibitor PsiB | - |
| VK940_RS24550 (VK940_24550) | 63631..64393 | + | 763 | Protein_69 | plasmid SOS inhibition protein A | - |
| VK940_RS24555 (VK940_24555) | 64567..64716 | + | 150 | Protein_70 | DUF5431 family protein | - |
| VK940_RS24560 (VK940_24560) | 64658..64783 | + | 126 | WP_231822392 | type I toxin-antitoxin system Hok family toxin | - |
| VK940_RS24565 (VK940_24565) | 65084..65380 | - | 297 | Protein_72 | hypothetical protein | - |
| VK940_RS24570 (VK940_24570) | 65450..65656 | + | 207 | WP_000547965 | hypothetical protein | - |
| VK940_RS24575 (VK940_24575) | 65681..65968 | + | 288 | WP_000107542 | hypothetical protein | - |
| VK940_RS24580 (VK940_24580) | 66087..66908 | + | 822 | WP_324649237 | DUF932 domain-containing protein | - |
| VK940_RS24585 (VK940_24585) | 67204..67794 | - | 591 | WP_001375132 | transglycosylase SLT domain-containing protein | virB1 |
| VK940_RS24590 (VK940_24590) | 68150..68533 | + | 384 | WP_001063020 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| VK940_RS24595 (VK940_24595) | 68724..69370 | + | 647 | Protein_78 | conjugal transfer transcriptional regulator TraJ | - |
| VK940_RS24600 (VK940_24600) | 69506..69721 | + | 216 | WP_024226173 | conjugal transfer relaxosome protein TraY | - |
| VK940_RS24605 (VK940_24605) | 69764..70129 | + | 366 | WP_000340282 | type IV conjugative transfer system pilin TraA | - |
| VK940_RS24610 (VK940_24610) | 70144..70455 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| VK940_RS24615 (VK940_24615) | 70477..71043 | + | 567 | WP_096306498 | type IV conjugative transfer system protein TraE | traE |
| VK940_RS24620 (VK940_24620) | 71030..71758 | + | 729 | WP_024226175 | type-F conjugative transfer system secretin TraK | traK |
| VK940_RS24625 (VK940_24625) | 71758..73185 | + | 1428 | WP_324649238 | F-type conjugal transfer pilus assembly protein TraB | traB |
| VK940_RS24630 (VK940_24630) | 73175..73765 | + | 591 | WP_324649239 | conjugal transfer pilus-stabilizing protein TraP | - |
| VK940_RS24635 (VK940_24635) | 73752..73949 | + | 198 | WP_024226177 | conjugal transfer protein TrbD | - |
| VK940_RS24640 (VK940_24640) | 73961..74212 | + | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| VK940_RS24645 (VK940_24645) | 74209..74724 | + | 516 | WP_024226179 | type IV conjugative transfer system lipoprotein TraV | traV |
| VK940_RS24650 (VK940_24650) | 74859..75080 | + | 222 | WP_001278692 | conjugal transfer protein TraR | - |
| VK940_RS24655 (VK940_24655) | 75240..77867 | + | 2628 | WP_024226180 | type IV secretion system protein TraC | virb4 |
| VK940_RS24660 (VK940_24660) | 77864..78250 | + | 387 | WP_000099690 | type-F conjugative transfer system protein TrbI | - |
| VK940_RS24665 (VK940_24665) | 78247..78879 | + | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| VK940_RS24670 (VK940_24670) | 78876..79868 | + | 993 | WP_024226181 | conjugal transfer pilus assembly protein TraU | traU |
| VK940_RS24675 (VK940_24675) | 79877..80515 | + | 639 | WP_324649244 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| VK940_RS24680 (VK940_24680) | 80512..82320 | + | 1809 | WP_097412160 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| VK940_RS24685 (VK940_24685) | 82347..82559 | + | 213 | WP_029785264 | conjugal transfer protein TrbE | - |
| VK940_RS24690 (VK940_24690) | 82597..83340 | + | 744 | WP_024226183 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| VK940_RS24695 (VK940_24695) | 83356..83703 | + | 348 | WP_024226184 | conjugal transfer protein TrbA | - |
| VK940_RS24700 (VK940_24700) | 83822..84106 | + | 285 | WP_096245929 | type-F conjugative transfer system pilin chaperone TraQ | - |
| VK940_RS24705 (VK940_24705) | 84093..84638 | + | 546 | WP_016236310 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| VK940_RS24710 (VK940_24710) | 84568..84930 | + | 363 | WP_071587080 | P-type conjugative transfer protein TrbJ | - |
| VK940_RS24715 (VK940_24715) | 84930..86303 | + | 1374 | WP_000944322 | conjugal transfer pilus assembly protein TraH | traH |
| VK940_RS24720 (VK940_24720) | 86300..89122 | + | 2823 | WP_262930124 | conjugal transfer mating-pair stabilization protein TraG | traG |
| VK940_RS24725 (VK940_24725) | 89137..89622 | + | 486 | WP_324649240 | surface exclusion protein | - |
| VK940_RS24730 (VK940_24730) | 89685..90847 | + | 1163 | WP_085947598 | IS3-like element IS3 family transposase | - |
| VK940_RS24735 (VK940_24735) | 90933..91664 | + | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| VK940_RS24740 (VK940_24740) | 92064..93038 | - | 975 | WP_012478345 | IS30-like element ISApl1 family transposase | - |
Host bacterium
| ID | 20623 | GenBank | NZ_CP141902 |
| Plasmid name | 6S-65-1|p1 | Incompatibility group | IncFIB |
| Plasmid size | 180516 bp | Coordinate of oriT [Strand] | 67772..67862 [-] |
| Host baterium | Escherichia albertii strain 6S-65-1 |
Cargo genes
| Drug resistance gene | tet(A), sitABCD |
| Virulence gene | iucA, iucB, iucC, iutA |
| Metal resistance gene | merP, merT, merR, pcoD, pcoC, pcoB, pcoA, silP, silA, silB, silF, silC, silR, silS, silE, yfeC |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |