Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120068 |
| Name | oriT_pKqq_SB1124_2 |
| Organism | Klebsiella quasipneumoniae subsp. quasipneumoniae strain SB1124 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP084808 (104479..104527 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pKqq_SB1124_2
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file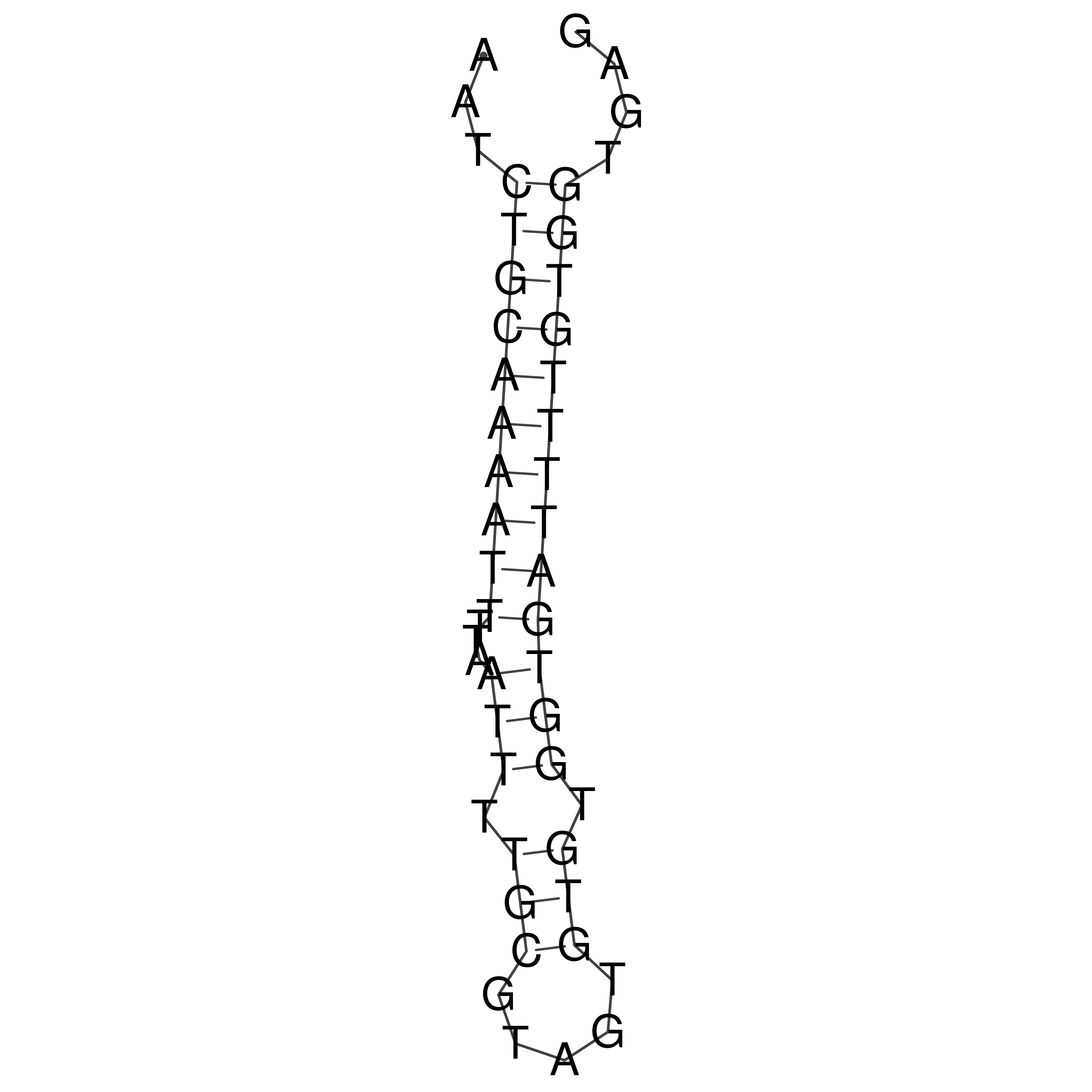
Relaxase
| ID | 12639 | GenBank | WP_204364702 |
| Name | traI_LGM22_RS27320_pKqq_SB1124_2 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190528.24 Da Isoelectric Point: 6.0252
>WP_204364702.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARAGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMESWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTIQ
AQIWSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIKK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARAGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMESWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTIQ
AQIWSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIKK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7133 | GenBank | WP_087829209 |
| Name | WP_087829209_pKqq_SB1124_2 |
UniProt ID | _ |
| Length | 129 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 129 a.a. Molecular weight: 14907.97 Da Isoelectric Point: 4.5248
>WP_087829209.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacteriaceae]
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEEE
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14840 | GenBank | WP_087829319 |
| Name | traD_LGM22_RS27315_pKqq_SB1124_2 |
UniProt ID | _ |
| Length | 761 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 761 a.a. Molecular weight: 85294.30 Da Isoelectric Point: 4.9677
>WP_087829319.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFIFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRSLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPAEVTV
SPAPVKATQTTKMPAEEPSVRATEPPVLRVTTVPLIKPKAAGTAATASSAGTPVAPAGGTEQELVQHSTE
QGRDMLPAGMNEDGVIEDMQAYDAWLADEQTLRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFIFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRSLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPAEVTV
SPAPVKATQTTKMPAEEPSVRATEPPVLRVTTVPLIKPKAAGTAATASSAGTPVAPAGGTEQELVQHSTE
QGRDMLPAGMNEDGVIEDMQAYDAWLADEQTLRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 103921..133751
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LGM22_RS27090 (LGM22_27090) | 99023..99373 | + | 351 | WP_004153414 | hypothetical protein | - |
| LGM22_RS27095 (LGM22_27095) | 100003..100356 | + | 354 | WP_004152748 | hypothetical protein | - |
| LGM22_RS27100 (LGM22_27100) | 100413..100760 | + | 348 | WP_004152749 | hypothetical protein | - |
| LGM22_RS27105 (LGM22_27105) | 100855..101001 | + | 147 | WP_004152750 | hypothetical protein | - |
| LGM22_RS27110 (LGM22_27110) | 101052..101885 | + | 834 | WP_004152751 | N-6 DNA methylase | - |
| LGM22_RS27115 (LGM22_27115) | 102705..103526 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| LGM22_RS27120 (LGM22_27120) | 103559..103888 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| LGM22_RS27125 (LGM22_27125) | 103921..104406 | - | 486 | WP_226683657 | transglycosylase SLT domain-containing protein | virB1 |
| LGM22_RS27130 (LGM22_27130) | 104839..105228 | + | 390 | WP_087829209 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LGM22_RS27135 (LGM22_27135) | 105467..106141 | + | 675 | WP_095033684 | hypothetical protein | - |
| LGM22_RS27140 (LGM22_27140) | 106333..106497 | + | 165 | WP_032444506 | TraY domain-containing protein | - |
| LGM22_RS27145 (LGM22_27145) | 106559..106927 | + | 369 | WP_087829211 | type IV conjugative transfer system pilin TraA | - |
| LGM22_RS27150 (LGM22_27150) | 106941..107246 | + | 306 | WP_087829212 | type IV conjugative transfer system protein TraL | traL |
| LGM22_RS27155 (LGM22_27155) | 107266..107832 | + | 567 | WP_064373744 | type IV conjugative transfer system protein TraE | traE |
| LGM22_RS27160 (LGM22_27160) | 107819..108559 | + | 741 | WP_020804840 | type-F conjugative transfer system secretin TraK | traK |
| LGM22_RS27165 (LGM22_27165) | 108559..109983 | + | 1425 | WP_023316397 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LGM22_RS27170 (LGM22_27170) | 109976..110572 | + | 597 | WP_023316398 | conjugal transfer pilus-stabilizing protein TraP | - |
| LGM22_RS27175 (LGM22_27175) | 110595..110864 | + | 270 | Protein_122 | type IV conjugative transfer system lipoprotein TraV | - |
| LGM22_RS27180 (LGM22_27180) | 110936..111853 | - | 918 | WP_028016400 | IS5-like element ISEc35 family transposase | - |
| LGM22_RS27185 (LGM22_27185) | 112045..112224 | + | 180 | Protein_124 | type IV conjugative transfer system lipoprotein TraV | - |
| LGM22_RS27190 (LGM22_27190) | 112356..112766 | + | 411 | WP_023316400 | hypothetical protein | - |
| LGM22_RS27195 (LGM22_27195) | 112771..113055 | + | 285 | WP_023316401 | hypothetical protein | - |
| LGM22_RS27200 (LGM22_27200) | 113079..113297 | + | 219 | WP_023316402 | hypothetical protein | - |
| LGM22_RS27205 (LGM22_27205) | 113298..113609 | + | 312 | WP_023316403 | hypothetical protein | - |
| LGM22_RS27210 (LGM22_27210) | 113676..114080 | + | 405 | WP_023316404 | hypothetical protein | - |
| LGM22_RS27215 (LGM22_27215) | 114077..114367 | + | 291 | WP_020323516 | hypothetical protein | - |
| LGM22_RS27220 (LGM22_27220) | 114375..114773 | + | 399 | WP_072156904 | hypothetical protein | - |
| LGM22_RS27225 (LGM22_27225) | 114845..117484 | + | 2640 | WP_020323518 | type IV secretion system protein TraC | virb4 |
| LGM22_RS27230 (LGM22_27230) | 117484..117873 | + | 390 | WP_020314627 | type-F conjugative transfer system protein TrbI | - |
| LGM22_RS27235 (LGM22_27235) | 117873..118499 | + | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| LGM22_RS27240 (LGM22_27240) | 118543..119502 | + | 960 | WP_029497356 | conjugal transfer pilus assembly protein TraU | traU |
| LGM22_RS27245 (LGM22_27245) | 119515..120153 | + | 639 | WP_064182677 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LGM22_RS27250 (LGM22_27250) | 120212..122167 | + | 1956 | WP_020803160 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LGM22_RS27255 (LGM22_27255) | 122200..122481 | + | 282 | WP_022644736 | hypothetical protein | - |
| LGM22_RS27260 (LGM22_27260) | 122471..122698 | + | 228 | WP_012540032 | conjugal transfer protein TrbE | - |
| LGM22_RS27265 (LGM22_27265) | 122709..123035 | + | 327 | WP_020323521 | hypothetical protein | - |
| LGM22_RS27270 (LGM22_27270) | 123056..123808 | + | 753 | WP_023316406 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LGM22_RS27275 (LGM22_27275) | 124263..124502 | + | 240 | WP_023316407 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LGM22_RS27280 (LGM22_27280) | 124474..125031 | + | 558 | WP_020804677 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LGM22_RS27285 (LGM22_27285) | 125077..125520 | + | 444 | WP_020804675 | F-type conjugal transfer protein TrbF | - |
| LGM22_RS27290 (LGM22_27290) | 125498..126877 | + | 1380 | WP_162495503 | conjugal transfer pilus assembly protein TraH | traH |
| LGM22_RS27295 (LGM22_27295) | 126877..129726 | + | 2850 | WP_032744228 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LGM22_RS27300 (LGM22_27300) | 129732..130259 | + | 528 | WP_020803849 | hypothetical protein | - |
| LGM22_RS27305 (LGM22_27305) | 130442..131110 | + | 669 | WP_020803846 | hypothetical protein | - |
| LGM22_RS27310 (LGM22_27310) | 131134..131394 | + | 261 | WP_020803850 | hypothetical protein | - |
| LGM22_RS27315 (LGM22_27315) | 131466..133751 | + | 2286 | WP_087829319 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 20498 | GenBank | NZ_CP084808 |
| Plasmid name | pKqq_SB1124_2 | Incompatibility group | IncFIB |
| Plasmid size | 144916 bp | Coordinate of oriT [Strand] | 104479..104527 [-] |
| Host baterium | Klebsiella quasipneumoniae subsp. quasipneumoniae strain SB1124 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merC, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |