Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120058 |
| Name | oriT_pKqq_SB98_2 |
| Organism | Klebsiella quasipneumoniae subsp. quasipneumoniae strain SB98 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP084805 (160884..160933 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pKqq_SB98_2
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file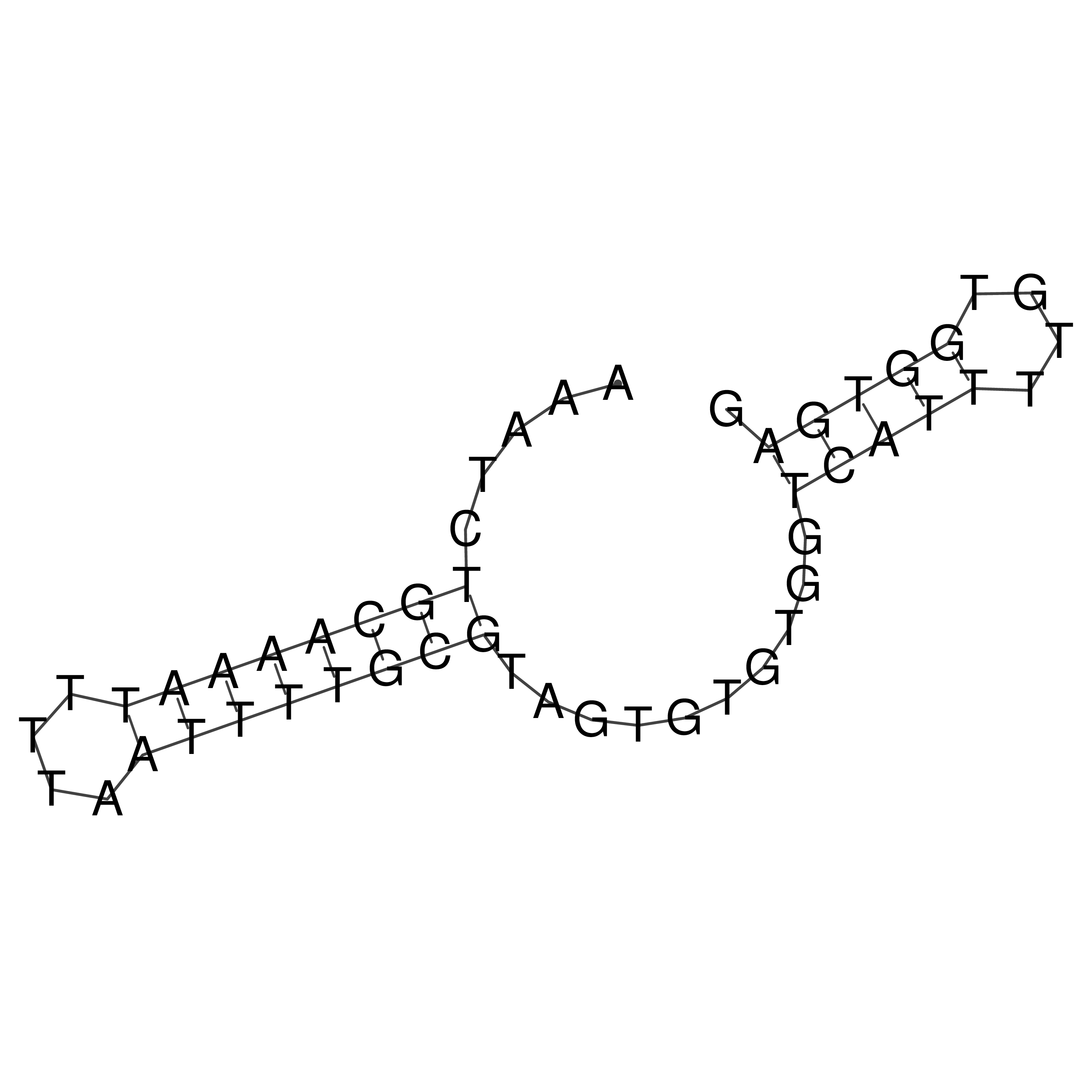
Relaxase
| ID | 12635 | GenBank | WP_016831059 |
| Name | traI_LGM23_RS28400_pKqq_SB98_2 |
UniProt ID | A0A483GHZ7 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190726.62 Da Isoelectric Point: 6.1403
>WP_016831059.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELLEAAGPDASLKSRDVAALDTRKSKEAIDPAENMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETAMVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRIWAEPAPVAPVPQTGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELLEAAGPDASLKSRDVAALDTRKSKEAIDPAENMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETAMVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRIWAEPAPVAPVPQTGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14835 | GenBank | WP_016831058 |
| Name | traD_LGM23_RS28405_pKqq_SB98_2 |
UniProt ID | _ |
| Length | 760 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 760 a.a. Molecular weight: 84900.95 Da Isoelectric Point: 5.0541
>WP_016831058.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSPPA
PAEMTVSPAPVKAPPTTKRPAAEPPVLPVTPIPLLKPKAAAAAATASSAGTPAAAAGGTEQELAQQSAEQ
GQDMLPAGMNEDGVIEDMQAYAAWLADDQTQRDMQRREEVNINHNHRHDEQDDVEIGGNI
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSPPA
PAEMTVSPAPVKAPPTTKRPAAEPPVLPVTPIPLLKPKAAAAAATASSAGTPAAAAGGTEQELAQQSAEQ
GQDMLPAGMNEDGVIEDMQAYAAWLADDQTQRDMQRREEVNINHNHRHDEQDDVEIGGNI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 131496..161491
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LGM23_RS28405 (LGM23_28405) | 131496..133778 | - | 2283 | WP_016831058 | type IV conjugative transfer system coupling protein TraD | virb4 |
| LGM23_RS28410 (LGM23_28410) | 133905..134594 | - | 690 | WP_072159541 | hypothetical protein | - |
| LGM23_RS28415 (LGM23_28415) | 134787..135518 | - | 732 | WP_016831056 | conjugal transfer complement resistance protein TraT | - |
| LGM23_RS28420 (LGM23_28420) | 135769..136371 | - | 603 | WP_071604857 | hypothetical protein | - |
| LGM23_RS28425 (LGM23_28425) | 136394..139216 | - | 2823 | WP_016831055 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LGM23_RS28430 (LGM23_28430) | 139216..140595 | - | 1380 | WP_073549766 | conjugal transfer pilus assembly protein TraH | traH |
| LGM23_RS28435 (LGM23_28435) | 140573..141016 | - | 444 | WP_016831053 | F-type conjugal transfer protein TrbF | - |
| LGM23_RS28440 (LGM23_28440) | 141062..141619 | - | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LGM23_RS28445 (LGM23_28445) | 141591..141830 | - | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LGM23_RS28450 (LGM23_28450) | 141841..142593 | - | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LGM23_RS28455 (LGM23_28455) | 142614..142940 | - | 327 | WP_016831051 | hypothetical protein | - |
| LGM23_RS28460 (LGM23_28460) | 142953..143201 | - | 249 | WP_004152675 | hypothetical protein | - |
| LGM23_RS28465 (LGM23_28465) | 143179..143433 | - | 255 | WP_015065558 | conjugal transfer protein TrbE | - |
| LGM23_RS28470 (LGM23_28470) | 143465..145420 | - | 1956 | WP_016831050 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LGM23_RS28475 (LGM23_28475) | 145479..146126 | - | 648 | WP_016831049 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LGM23_RS28480 (LGM23_28480) | 146205..146381 | - | 177 | WP_016831048 | hypothetical protein | - |
| LGM23_RS28485 (LGM23_28485) | 146402..147391 | - | 990 | WP_015632509 | conjugal transfer pilus assembly protein TraU | traU |
| LGM23_RS28490 (LGM23_28490) | 147388..147789 | - | 402 | WP_016831047 | hypothetical protein | - |
| LGM23_RS28495 (LGM23_28495) | 147824..148459 | - | 636 | WP_016831046 | type-F conjugative transfer system protein TraW | traW |
| LGM23_RS28500 (LGM23_28500) | 148459..148848 | - | 390 | WP_004167468 | type-F conjugative transfer system protein TrbI | - |
| LGM23_RS28505 (LGM23_28505) | 148848..151487 | - | 2640 | WP_016831045 | type IV secretion system protein TraC | virb4 |
| LGM23_RS28510 (LGM23_28510) | 151559..151957 | - | 399 | WP_023158008 | hypothetical protein | - |
| LGM23_RS28515 (LGM23_28515) | 151965..152183 | - | 219 | WP_072196435 | hypothetical protein | - |
| LGM23_RS28520 (LGM23_28520) | 152197..152355 | - | 159 | WP_223338907 | hypothetical protein | - |
| LGM23_RS28525 (LGM23_28525) | 152398..152802 | - | 405 | WP_016831042 | hypothetical protein | - |
| LGM23_RS28530 (LGM23_28530) | 152869..153180 | - | 312 | WP_016831041 | hypothetical protein | - |
| LGM23_RS28535 (LGM23_28535) | 153181..153399 | - | 219 | WP_014386199 | hypothetical protein | - |
| LGM23_RS28540 (LGM23_28540) | 153423..153712 | - | 290 | Protein_161 | hypothetical protein | - |
| LGM23_RS28545 (LGM23_28545) | 153717..154127 | - | 411 | WP_016831039 | hypothetical protein | - |
| LGM23_RS28550 (LGM23_28550) | 154259..154843 | - | 585 | WP_014386196 | type IV conjugative transfer system lipoprotein TraV | traV |
| LGM23_RS28555 (LGM23_28555) | 154866..155462 | - | 597 | Protein_164 | conjugal transfer pilus-stabilizing protein TraP | - |
| LGM23_RS28560 (LGM23_28560) | 155455..156879 | - | 1425 | WP_016831036 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LGM23_RS28565 (LGM23_28565) | 156879..157619 | - | 741 | WP_014386193 | type-F conjugative transfer system secretin TraK | traK |
| LGM23_RS28570 (LGM23_28570) | 157606..158172 | - | 567 | WP_016831035 | type IV conjugative transfer system protein TraE | traE |
| LGM23_RS28575 (LGM23_28575) | 158192..158497 | - | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| LGM23_RS28580 (LGM23_28580) | 158511..158879 | - | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| LGM23_RS28585 (LGM23_28585) | 158941..159147 | - | 207 | WP_171773970 | TraY domain-containing protein | - |
| LGM23_RS28590 (LGM23_28590) | 159280..159999 | - | 720 | WP_016831034 | conjugal transfer protein TrbJ | - |
| LGM23_RS28595 (LGM23_28595) | 160199..160615 | - | 417 | WP_072145360 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LGM23_RS28600 (LGM23_28600) | 161006..161491 | + | 486 | WP_004178063 | transglycosylase SLT domain-containing protein | virB1 |
| LGM23_RS28605 (LGM23_28605) | 161524..161853 | - | 330 | WP_011977736 | DUF5983 family protein | - |
| LGM23_RS28610 (LGM23_28610) | 161886..162707 | - | 822 | WP_004182076 | DUF932 domain-containing protein | - |
| LGM23_RS28615 (LGM23_28615) | 163540..163953 | - | 414 | WP_004182074 | type II toxin-antitoxin system HigA family antitoxin | - |
| LGM23_RS28620 (LGM23_28620) | 163954..164232 | - | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| LGM23_RS28625 (LGM23_28625) | 164222..164542 | - | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| LGM23_RS28630 (LGM23_28630) | 164623..164847 | - | 225 | WP_004152719 | hypothetical protein | - |
| LGM23_RS28635 (LGM23_28635) | 164858..165070 | - | 213 | WP_004182070 | hypothetical protein | - |
| LGM23_RS28640 (LGM23_28640) | 165131..165487 | - | 357 | WP_004152717 | hypothetical protein | - |
| LGM23_RS28645 (LGM23_28645) | 166123..166473 | - | 351 | WP_004182068 | hypothetical protein | - |
Host bacterium
| ID | 20488 | GenBank | NZ_CP084805 |
| Plasmid name | pKqq_SB98_2 | Incompatibility group | IncFIB |
| Plasmid size | 195131 bp | Coordinate of oriT [Strand] | 160884..160933 [+] |
| Host baterium | Klebsiella quasipneumoniae subsp. quasipneumoniae strain SB98 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE, arsH, arsC, arsB, arsA, arsD, arsR, fecD, fecE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |