Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120015 |
| Name | oriT_pKqs_CIP110288_1 |
| Organism | Klebsiella quasipneumoniae subsp. similipneumoniae strain CIP110288 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP084778 (112494..112543 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pKqs_CIP110288_1
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file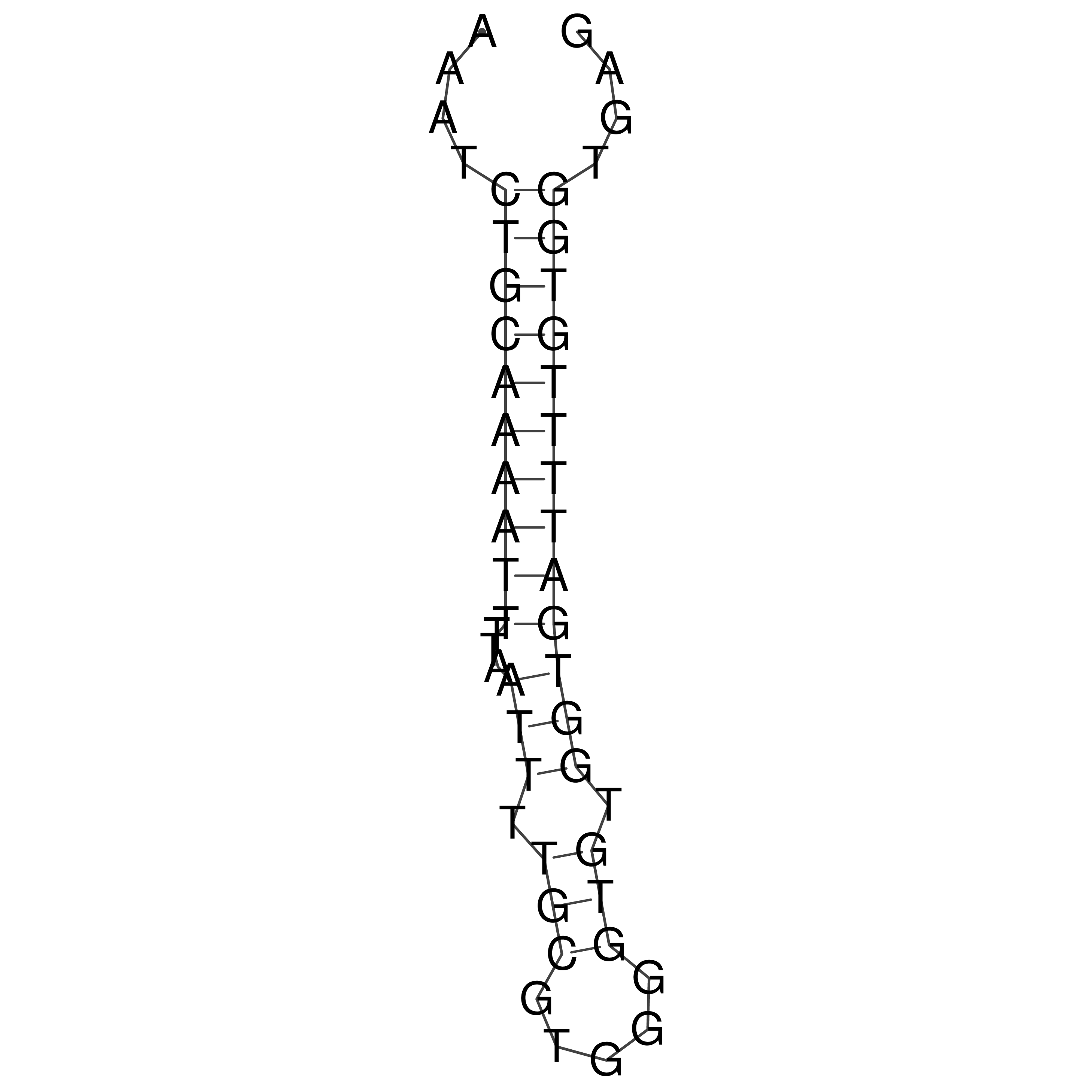
Relaxase
| ID | 12617 | GenBank | WP_063106403 |
| Name | traI_LGM28_RS25630_pKqs_CIP110288_1 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190775.71 Da Isoelectric Point: 6.0387
>WP_063106403.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKTLFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQTLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKD
TTITSLTPVWLDSKSRGVRDYYREGMVMERWDPENQTHDRFVIDRVTASSNMLTLKDREGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESIAVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLHTPAEQAIHLSIPLLESQDLTFTRPQLLATALETGGDKVPMWSIEMTI
KKQIKSGQLLNVGVSPGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPHVIGLGPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSISPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDIDRALATIEQVTPERVPRKEGAWVPGSSVVEFTPTQEEEIR
KALSKGESLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNNDRRALNGLIHEARRENGETGREEVTL
PVLVTSNIRDGELRKLSTWTAHKGAVALVDNVYHRIDNVDKDNQLITLTDSEGKERYISPREASAEGVTL
YRQEEITVSQGDRMRFSKSDPERGYVANSVWEVQSVSGDSVTLSDGKTTRTLTPKADEAQQHIDLAYAIT
AHGAQGASEPYAIALEGMVGARKLMASFESAYVGLSRMKQHVQVYTDRREGWIKAIQHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNIGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAITGGRVWAEQAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
VERVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASETLQRDRLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKTLFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQTLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKD
TTITSLTPVWLDSKSRGVRDYYREGMVMERWDPENQTHDRFVIDRVTASSNMLTLKDREGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESIAVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLHTPAEQAIHLSIPLLESQDLTFTRPQLLATALETGGDKVPMWSIEMTI
KKQIKSGQLLNVGVSPGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPHVIGLGPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSISPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDIDRALATIEQVTPERVPRKEGAWVPGSSVVEFTPTQEEEIR
KALSKGESLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNNDRRALNGLIHEARRENGETGREEVTL
PVLVTSNIRDGELRKLSTWTAHKGAVALVDNVYHRIDNVDKDNQLITLTDSEGKERYISPREASAEGVTL
YRQEEITVSQGDRMRFSKSDPERGYVANSVWEVQSVSGDSVTLSDGKTTRTLTPKADEAQQHIDLAYAIT
AHGAQGASEPYAIALEGMVGARKLMASFESAYVGLSRMKQHVQVYTDRREGWIKAIQHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNIGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAITGGRVWAEQAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
VERVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASETLQRDRLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14815 | GenBank | WP_063106402 |
| Name | traD_LGM28_RS25635_pKqs_CIP110288_1 |
UniProt ID | _ |
| Length | 768 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 768 a.a. Molecular weight: 85829.88 Da Isoelectric Point: 5.0689
>WP_063106402.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPIGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSQPA
PAEVTVSPAPVKAPPTTKSPAAEPSARAAEPPVLRVTTVPLIKPKAAAAAAAAATASSVGTPAAGGTEQE
LAQQSAEQGQEMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINRSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPIGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSQPA
PAEVTVSPAPVKAPPTTKSPAAEPSARAAEPPVLRVTTVPLIKPKAAAAAAAAATASSVGTPAAGGTEQE
LAQQSAEQGQEMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINRSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 84049..109996
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LGM28_RS25635 (LGM28_25635) | 84049..86355 | - | 2307 | WP_063106402 | type IV conjugative transfer system coupling protein TraD | virb4 |
| LGM28_RS25640 (LGM28_25640) | 86429..86524 | - | 96 | Protein_69 | hypothetical protein | - |
| LGM28_RS25645 (LGM28_25645) | 86717..87448 | - | 732 | WP_004152629 | conjugal transfer complement resistance protein TraT | - |
| LGM28_RS25650 (LGM28_25650) | 87638..88165 | - | 528 | WP_136084787 | conjugal transfer protein TraS | - |
| LGM28_RS25655 (LGM28_25655) | 88171..91020 | - | 2850 | WP_136084783 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LGM28_RS25660 (LGM28_25660) | 91020..92390 | - | 1371 | WP_047665852 | conjugal transfer pilus assembly protein TraH | traH |
| LGM28_RS25665 (LGM28_25665) | 92377..92799 | - | 423 | WP_049010971 | hypothetical protein | - |
| LGM28_RS25670 (LGM28_25670) | 92792..93364 | - | 573 | WP_055323574 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LGM28_RS25675 (LGM28_25675) | 93336..93575 | - | 240 | WP_063106400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LGM28_RS25680 (LGM28_25680) | 93586..94338 | - | 753 | WP_136084782 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LGM28_RS25685 (LGM28_25685) | 94359..94685 | - | 327 | WP_012539967 | hypothetical protein | - |
| LGM28_RS25690 (LGM28_25690) | 94696..94923 | - | 228 | WP_047665846 | conjugal transfer protein TrbE | - |
| LGM28_RS25695 (LGM28_25695) | 94913..95194 | - | 282 | WP_047665844 | hypothetical protein | - |
| LGM28_RS25700 (LGM28_25700) | 95227..97182 | - | 1956 | WP_063106399 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LGM28_RS25705 (LGM28_25705) | 97241..97879 | - | 639 | WP_047667531 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LGM28_RS25710 (LGM28_25710) | 97892..98881 | - | 990 | WP_046622837 | conjugal transfer pilus assembly protein TraU | traU |
| LGM28_RS25715 (LGM28_25715) | 98878..99267 | - | 390 | WP_047667530 | hypothetical protein | - |
| LGM28_RS25720 (LGM28_25720) | 99309..99935 | - | 627 | WP_004152507 | type-F conjugative transfer system protein TraW | traW |
| LGM28_RS25725 (LGM28_25725) | 99935..100324 | - | 390 | WP_136084781 | type-F conjugative transfer system protein TrbI | - |
| LGM28_RS25730 (LGM28_25730) | 100324..102963 | - | 2640 | WP_046622834 | type IV secretion system protein TraC | virb4 |
| LGM28_RS25735 (LGM28_25735) | 103035..103433 | - | 399 | WP_168429906 | hypothetical protein | - |
| LGM28_RS25740 (LGM28_25740) | 103441..103683 | - | 243 | WP_071885119 | hypothetical protein | - |
| LGM28_RS25745 (LGM28_25745) | 103875..104279 | - | 405 | WP_046622833 | hypothetical protein | - |
| LGM28_RS25750 (LGM28_25750) | 104346..104657 | - | 312 | Protein_91 | hypothetical protein | - |
| LGM28_RS25755 (LGM28_25755) | 104658..104876 | - | 219 | WP_014386199 | hypothetical protein | - |
| LGM28_RS25760 (LGM28_25760) | 104900..105190 | - | 291 | WP_047665765 | hypothetical protein | - |
| LGM28_RS25765 (LGM28_25765) | 105219..105617 | - | 399 | WP_223170083 | hypothetical protein | - |
| LGM28_RS25770 (LGM28_25770) | 105773..106342 | - | 570 | WP_046622829 | type IV conjugative transfer system lipoprotein TraV | traV |
| LGM28_RS25775 (LGM28_25775) | 106365..106961 | - | 597 | WP_136084780 | conjugal transfer pilus-stabilizing protein TraP | - |
| LGM28_RS25780 (LGM28_25780) | 106954..108378 | - | 1425 | WP_136084779 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LGM28_RS25785 (LGM28_25785) | 108378..109118 | - | 741 | WP_063106395 | type-F conjugative transfer system secretin TraK | traK |
| LGM28_RS25790 (LGM28_25790) | 109105..109671 | - | 567 | WP_063106394 | type IV conjugative transfer system protein TraE | traE |
| LGM28_RS25795 (LGM28_25795) | 109691..109996 | - | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| LGM28_RS25800 (LGM28_25800) | 110010..110378 | - | 369 | WP_047665762 | type IV conjugative transfer system pilin TraA | - |
| LGM28_RS25805 (LGM28_25805) | 110432..110803 | - | 372 | WP_071885111 | TraY domain-containing protein | - |
| LGM28_RS25810 (LGM28_25810) | 110887..111582 | - | 696 | WP_072194100 | transcriptional regulator TraJ family protein | - |
| LGM28_RS25815 (LGM28_25815) | 111789..112181 | - | 393 | WP_136084778 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LGM28_RS25820 (LGM28_25820) | 112688..113095 | + | 408 | WP_223170089 | transglycosylase SLT domain-containing protein | - |
| LGM28_RS25825 (LGM28_25825) | 113133..113663 | - | 531 | Protein_106 | antirestriction protein | - |
Host bacterium
| ID | 20445 | GenBank | NZ_CP084778 |
| Plasmid name | pKqs_CIP110288_1 | Incompatibility group | IncFIB |
| Plasmid size | 151441 bp | Coordinate of oriT [Strand] | 112494..112543 [+] |
| Host baterium | Klebsiella quasipneumoniae subsp. similipneumoniae strain CIP110288 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsH, arsC, arsB, arsA, arsD, arsR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |