Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 120009 |
| Name | oriT_pNUITM-VR1_2 |
| Organism | Raoultella ornithinolytica strain NUITM-VR1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP025011 (226292..226341 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..13, 18..24 (GCAAAAT..ATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pNUITM-VR1_2
AAATCTGCAAAATATTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATATTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file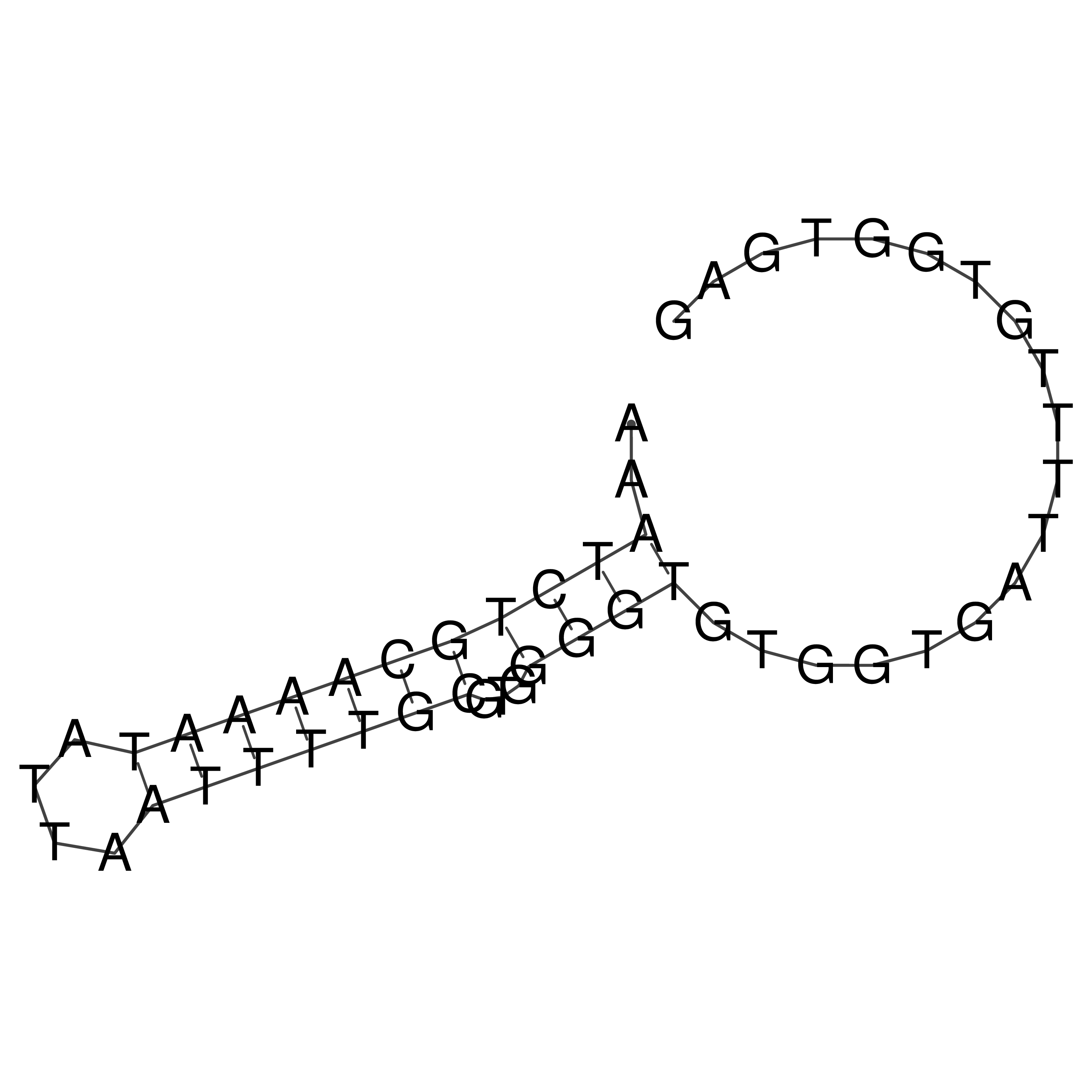
Relaxase
| ID | 12612 | GenBank | WP_224050193 |
| Name | traI_LDO46_RS29650_pNUITM-VR1_2 |
UniProt ID | _ |
| Length | 1754 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1754 a.a. Molecular weight: 190526.56 Da Isoelectric Point: 6.3185
>WP_224050193.1 conjugative transfer relaxase/helicase TraI [Raoultella ornithinolytica]
MMSIGSVKAAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTGSSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTSFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTQAQEKAIQ
KALSEGETLPAGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGSSPGRFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASEGLLQRERLQAVERDMVRDLNREKT
LGGD
MMSIGSVKAAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTGSSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTSFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTQAQEKAIQ
KALSEGETLPAGQPASLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGSSPGRFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASEGLLQRERLQAVERDMVRDLNREKT
LGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14810 | GenBank | WP_032454979 |
| Name | traD_LDO46_RS29655_pNUITM-VR1_2 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 86062.11 Da Isoelectric Point: 5.0175
>WP_032454979.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKRPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDEVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKRPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDEVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 198109..223976
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LDO46_RS29655 (NUITMVR1_58510) | 198109..200421 | - | 2313 | WP_032454979 | type IV conjugative transfer system coupling protein TraD | virb4 |
| LDO46_RS29660 (NUITMVR1_58520) | 200548..201237 | - | 690 | WP_013023831 | hypothetical protein | - |
| LDO46_RS29665 (NUITMVR1_58530) | 201430..202161 | - | 732 | WP_013023830 | conjugal transfer complement resistance protein TraT | - |
| LDO46_RS29670 (NUITMVR1_58540) | 202347..202880 | - | 534 | WP_014343486 | conjugal transfer protein TraS | - |
| LDO46_RS29675 (NUITMVR1_58560) | 202883..205740 | - | 2858 | Protein_198 | conjugal transfer mating-pair stabilization protein TraG | - |
| LDO46_RS29680 (NUITMVR1_58570) | 205740..207119 | - | 1380 | WP_074184099 | conjugal transfer pilus assembly protein TraH | traH |
| LDO46_RS29685 (NUITMVR1_58580) | 207097..207540 | - | 444 | WP_032454976 | F-type conjugal transfer protein TrbF | - |
| LDO46_RS29690 (NUITMVR1_58590) | 207586..208143 | - | 558 | WP_055316562 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LDO46_RS29695 (NUITMVR1_58600) | 208115..208354 | - | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LDO46_RS29700 (NUITMVR1_58610) | 208365..209117 | - | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LDO46_RS29705 (NUITMVR1_58620) | 209138..209464 | - | 327 | WP_004144402 | hypothetical protein | - |
| LDO46_RS29710 (NUITMVR1_58630) | 209477..209725 | - | 249 | WP_004152675 | hypothetical protein | - |
| LDO46_RS29715 (NUITMVR1_58640) | 209703..209957 | - | 255 | WP_224050194 | conjugal transfer protein TrbE | - |
| LDO46_RS29720 (NUITMVR1_58650) | 209989..211944 | - | 1956 | WP_055316764 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LDO46_RS29725 (NUITMVR1_58660) | 212003..212641 | - | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LDO46_RS29730 (NUITMVR1_58670) | 212654..213643 | - | 990 | WP_057193347 | conjugal transfer pilus assembly protein TraU | traU |
| LDO46_RS29735 (NUITMVR1_58680) | 213640..214041 | - | 402 | WP_004194979 | hypothetical protein | - |
| LDO46_RS29740 (NUITMVR1_58690) | 214076..214711 | - | 636 | WP_032441885 | type-F conjugative transfer system protein TraW | traW |
| LDO46_RS29745 (NUITMVR1_58700) | 214711..215100 | - | 390 | WP_004167468 | type-F conjugative transfer system protein TrbI | - |
| LDO46_RS29750 (NUITMVR1_58710) | 215097..217739 | - | 2643 | WP_032454974 | type IV secretion system protein TraC | virb4 |
| LDO46_RS29755 (NUITMVR1_58720) | 217811..218209 | - | 399 | WP_074184101 | hypothetical protein | - |
| LDO46_RS29760 (NUITMVR1_58730) | 218587..218991 | - | 405 | WP_004197817 | hypothetical protein | - |
| LDO46_RS29765 (NUITMVR1_58740) | 219058..219369 | - | 312 | WP_004195240 | hypothetical protein | - |
| LDO46_RS29770 (NUITMVR1_58750) | 219370..219588 | - | 219 | WP_004195235 | hypothetical protein | - |
| LDO46_RS29775 (NUITMVR1_58760) | 219694..220104 | - | 411 | WP_004152499 | hypothetical protein | - |
| LDO46_RS29780 (NUITMVR1_58770) | 220236..220820 | - | 585 | WP_032441881 | type IV conjugative transfer system lipoprotein TraV | traV |
| LDO46_RS29785 (NUITMVR1_58780) | 220934..222358 | - | 1425 | WP_023284634 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LDO46_RS29790 (NUITMVR1_58790) | 222358..223098 | - | 741 | WP_013214019 | type-F conjugative transfer system secretin TraK | traK |
| LDO46_RS29795 (NUITMVR1_58800) | 223085..223651 | - | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| LDO46_RS29800 (NUITMVR1_58810) | 223671..223976 | - | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| LDO46_RS29805 (NUITMVR1_58820) | 223990..224358 | - | 369 | WP_224050195 | type IV conjugative transfer system pilin TraA | - |
| LDO46_RS29810 (NUITMVR1_58830) | 224388..224567 | - | 180 | WP_224050196 | TraY domain-containing protein | - |
| LDO46_RS29815 (NUITMVR1_58840) | 224652..225353 | - | 702 | WP_032736872 | hypothetical protein | - |
| LDO46_RS29820 (NUITMVR1_58850) | 225592..225984 | - | 393 | WP_032736874 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LDO46_RS29825 (NUITMVR1_58860) | 226416..226892 | + | 477 | WP_076752132 | transglycosylase SLT domain-containing protein | - |
| LDO46_RS29830 (NUITMVR1_58870) | 226930..227460 | - | 531 | WP_076752134 | antirestriction protein | - |
| LDO46_RS29835 (NUITMVR1_58880) | 227652..227882 | + | 231 | WP_076752135 | hypothetical protein | - |
| LDO46_RS29840 (NUITMVR1_58890) | 228086..228919 | - | 834 | WP_076752137 | N-6 DNA methylase | - |
Host bacterium
| ID | 20439 | GenBank | NZ_AP025011 |
| Plasmid name | pNUITM-VR1_2 | Incompatibility group | IncFIB |
| Plasmid size | 261835 bp | Coordinate of oriT [Strand] | 226292..226341 [+] |
| Host baterium | Raoultella ornithinolytica strain NUITM-VR1 |
Cargo genes
| Drug resistance gene | mcr-10, qnrS1, aph(6)-Id, aph(3'')-Ib |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merC, merP, merT, merR, terW, terZ, terA, terB, terC, terD, terE, arsH, arsC, arsB, arsA, arsD, arsR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |