Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119995 |
| Name | oriT_FDAARGOS_1503|unnamed1 |
| Organism | Klebsiella quasipneumoniae strain FDAARGOS_1503 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP083634 (7472..7521 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_FDAARGOS_1503|unnamed1
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file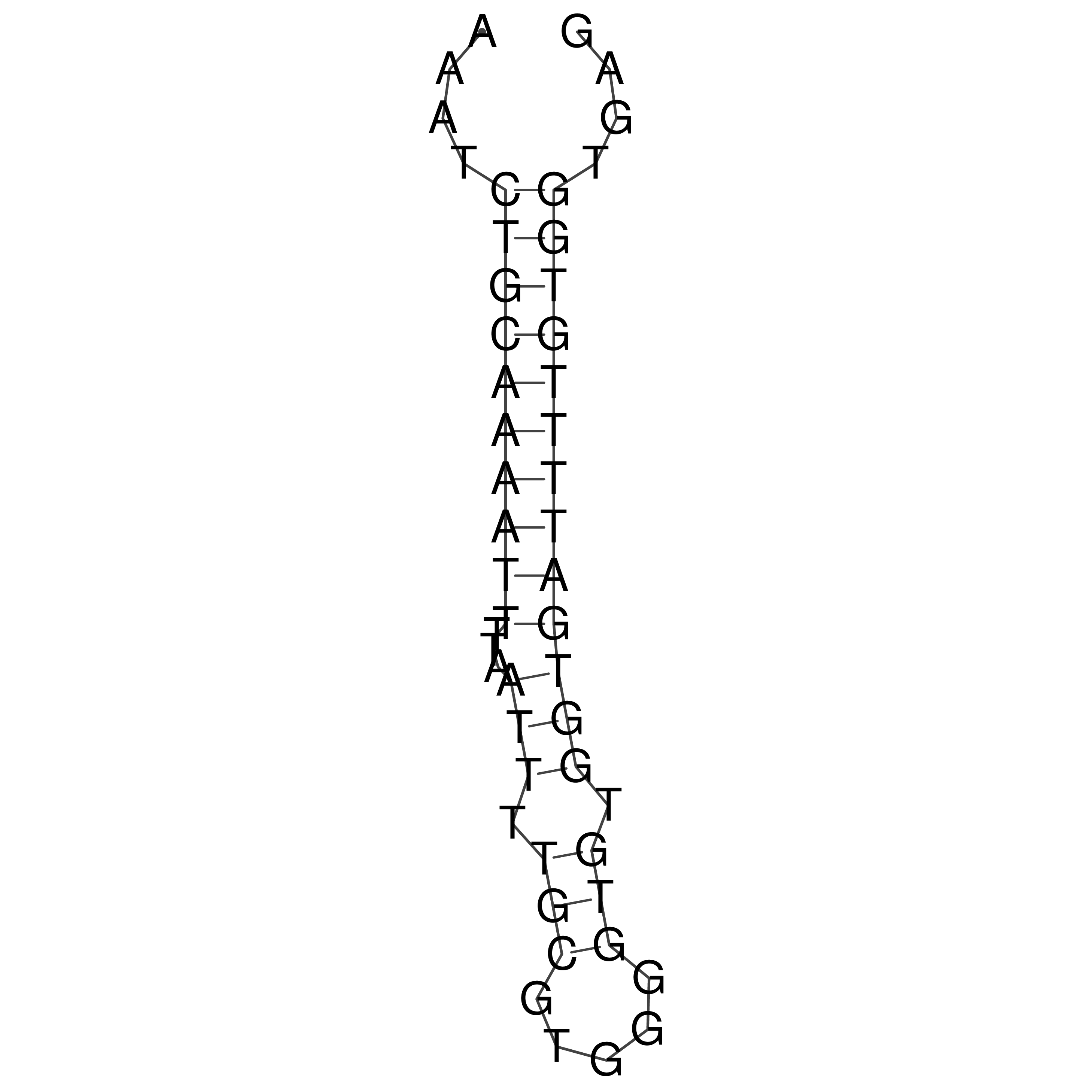
Relaxase
| ID | 12603 | GenBank | WP_044525168 |
| Name | traI_LA349_RS25795_FDAARGOS_1503|unnamed1 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190549.78 Da Isoelectric Point: 6.4837
>WP_044525168.1 conjugative transfer relaxase/helicase TraI [Klebsiella quasipneumoniae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDTDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALITIEQVTPEQVPRKEGVWAPGSSVVEFTPKQEKAIE
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRTLNGMIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSDGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASERYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRIWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDTDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALITIEQVTPEQVPRKEGVWAPGSSVVEFTPKQEKAIE
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRTLNGMIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSDGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASERYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRIWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14802 | GenBank | WP_015065643 |
| Name | traD_LA349_RS25790_FDAARGOS_1503|unnamed1 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85851.84 Da Isoelectric Point: 5.0577
>WP_015065643.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTVWCGEQLWTSIVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRVDARLSALLEAREAEGSLARTLFMPDAPASGPADTNSHAGEQPEPISQPA
PADMTVSPAPVKAPPTTKSPAAEPSVRATEPPVLRVTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTVWCGEQLWTSIVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRVDARLSALLEAREAEGSLARTLFMPDAPASGPADTNSHAGEQPEPISQPA
PADMTVSPAPVKAPPTTKSPAAEPSVRATEPPVLRVTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6914..35408
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LA349_RS25575 (LA349_25575) | 1934..2284 | + | 351 | WP_023280874 | hypothetical protein | - |
| LA349_RS25580 (LA349_25580) | 2921..3277 | + | 357 | WP_004152717 | hypothetical protein | - |
| LA349_RS25585 (LA349_25585) | 3338..3550 | + | 213 | WP_004152718 | hypothetical protein | - |
| LA349_RS25590 (LA349_25590) | 3561..3785 | + | 225 | WP_004152719 | hypothetical protein | - |
| LA349_RS25595 (LA349_25595) | 3866..4186 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| LA349_RS25600 (LA349_25600) | 4176..4454 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| LA349_RS25605 (LA349_25605) | 4455..4868 | + | 414 | WP_023280875 | type II toxin-antitoxin system HigA family antitoxin | - |
| LA349_RS25610 (LA349_25610) | 5698..6525 | + | 828 | WP_023307552 | DUF932 domain-containing protein | - |
| LA349_RS25615 (LA349_25615) | 6552..6881 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| LA349_RS25620 (LA349_25620) | 6914..7399 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| LA349_RS25625 (LA349_25625) | 7834..8226 | + | 393 | WP_004206766 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LA349_RS25630 (LA349_25630) | 8433..9128 | + | 696 | WP_023307551 | transcriptional regulator TraJ family protein | - |
| LA349_RS25635 (LA349_25635) | 9212..9583 | + | 372 | WP_004208838 | TraY domain-containing protein | - |
| LA349_RS25640 (LA349_25640) | 9637..10005 | + | 369 | WP_023307550 | type IV conjugative transfer system pilin TraA | - |
| LA349_RS25645 (LA349_25645) | 10019..10324 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| LA349_RS25650 (LA349_25650) | 10344..10910 | + | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| LA349_RS25655 (LA349_25655) | 10897..11637 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| LA349_RS25660 (LA349_25660) | 11637..13061 | + | 1425 | WP_023307549 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LA349_RS25665 (LA349_25665) | 13054..13467 | + | 414 | Protein_21 | conjugal transfer pilus-stabilizing protein TraP | - |
| LA349_RS25670 (LA349_25670) | 13679..14248 | + | 570 | WP_023307547 | type IV conjugative transfer system lipoprotein TraV | traV |
| LA349_RS25675 (LA349_25675) | 14380..14790 | + | 411 | WP_023307546 | hypothetical protein | - |
| LA349_RS25680 (LA349_25680) | 14795..15085 | + | 291 | WP_032423381 | hypothetical protein | - |
| LA349_RS25685 (LA349_25685) | 15109..15327 | + | 219 | WP_023307545 | hypothetical protein | - |
| LA349_RS25690 (LA349_25690) | 15328..15645 | + | 318 | WP_023307544 | hypothetical protein | - |
| LA349_RS25695 (LA349_25695) | 15712..16116 | + | 405 | WP_023307543 | hypothetical protein | - |
| LA349_RS25700 (LA349_25700) | 16113..16403 | + | 291 | WP_023307542 | hypothetical protein | - |
| LA349_RS25705 (LA349_25705) | 16411..16809 | + | 399 | WP_072158599 | hypothetical protein | - |
| LA349_RS25710 (LA349_25710) | 16881..19520 | + | 2640 | WP_016528865 | type IV secretion system protein TraC | virb4 |
| LA349_RS25715 (LA349_25715) | 19520..19921 | + | 402 | WP_023307539 | type-F conjugative transfer system protein TrbI | - |
| LA349_RS25720 (LA349_25720) | 19921..20547 | + | 627 | WP_023307538 | type-F conjugative transfer system protein TraW | traW |
| LA349_RS25725 (LA349_25725) | 20591..21550 | + | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| LA349_RS25730 (LA349_25730) | 21563..22201 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LA349_RS25735 (LA349_25735) | 22260..24215 | + | 1956 | WP_023307537 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LA349_RS25740 (LA349_25740) | 24247..24483 | + | 237 | WP_023307536 | conjugal transfer protein TrbE | - |
| LA349_RS26515 | 24480..24665 | + | 186 | WP_023307535 | hypothetical protein | - |
| LA349_RS25745 (LA349_25745) | 24711..25037 | + | 327 | WP_004152685 | hypothetical protein | - |
| LA349_RS25750 (LA349_25750) | 25058..25810 | + | 753 | WP_015065637 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LA349_RS25755 (LA349_25755) | 25821..26060 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LA349_RS25760 (LA349_25760) | 26032..26589 | + | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LA349_RS25765 (LA349_25765) | 26634..27077 | + | 444 | WP_015065638 | F-type conjugal transfer protein TrbF | - |
| LA349_RS25770 (LA349_25770) | 27055..28434 | + | 1380 | WP_011977731 | conjugal transfer pilus assembly protein TraH | traH |
| LA349_RS25775 (LA349_25775) | 28434..31283 | + | 2850 | WP_023307534 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LA349_RS25780 (LA349_25780) | 31289..31816 | + | 528 | WP_032423397 | conjugal transfer protein TraS | - |
| LA349_RS25785 (LA349_25785) | 32006..32737 | + | 732 | Protein_46 | conjugal transfer complement resistance protein TraT | - |
| LA349_RS25790 (LA349_25790) | 33099..35408 | + | 2310 | WP_015065643 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 20425 | GenBank | NZ_CP083634 |
| Plasmid name | FDAARGOS_1503|unnamed1 | Incompatibility group | IncFII |
| Plasmid size | 151102 bp | Coordinate of oriT [Strand] | 7472..7521 [-] |
| Host baterium | Klebsiella quasipneumoniae strain FDAARGOS_1503 |
Cargo genes
| Drug resistance gene | sul1, qacE, ant(3'')-Ia |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merF, merP, merT, merR2, arsC, arsB, fecD, fecE, arsR, arsH |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |