Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119622 |
| Name | oriT_SB18002|unnamed |
| Organism | Escherichia sp. SB18002 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP141235 (71437..71509 [-], 73 nt) |
| oriT length | 73 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 73 nt
>oriT_SB18002|unnamed
GTCGGGGCAAAGCCCTGACCAGGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
GTCGGGGCAAAGCCCTGACCAGGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file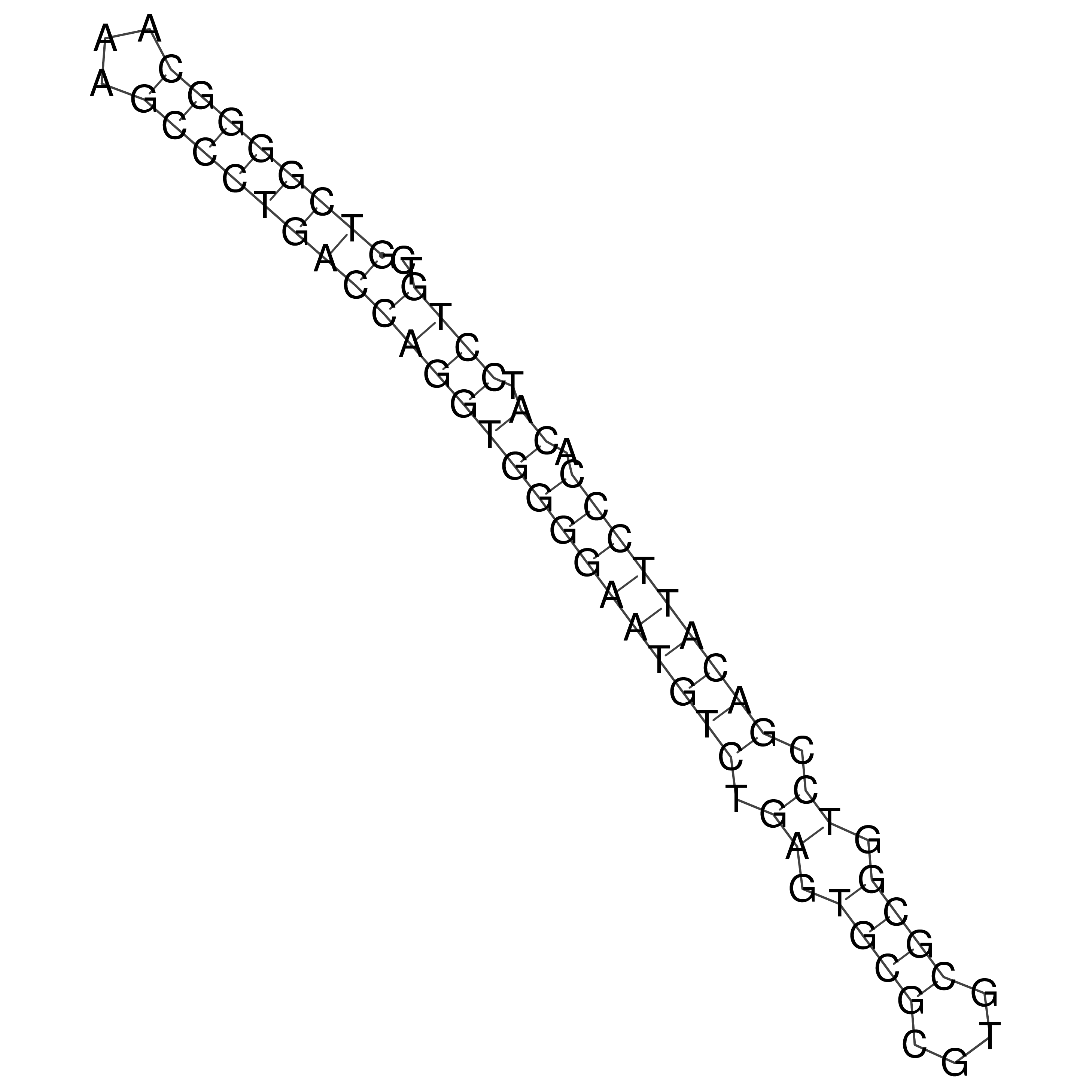
Relaxase
| ID | 12399 | GenBank | WP_029391987 |
| Name | Relaxase_VJF88_RS00435_SB18002|unnamed |
UniProt ID | A0A2H5C199 |
| Length | 906 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 105112.51 Da Isoelectric Point: 8.2682
>WP_029391987.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPEMARNGELESCLDVHRTLAKAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGHSIGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAGADIFERVKPECRYNPELAASDEVEPGFRRDPVLRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQVRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPEMARNGELESCLDVHRTLAKAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGHSIGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAGADIFERVKPECRYNPELAASDEVEPGFRRDPVLRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQVRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A2H5C199 |
Auxiliary protein
| ID | 7055 | GenBank | WP_001291058 |
| Name | WP_001291058_SB18002|unnamed |
UniProt ID | A0A7I0L241 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12730.52 Da Isoelectric Point: 9.9656
>WP_001291058.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7I0L241 |
T4CP
| ID | 14557 | GenBank | WP_029392471 |
| Name | trbC_VJF88_RS00440_SB18002|unnamed |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87239.20 Da Isoelectric Point: 6.6676
>WP_029392471.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSQHHVNPDLIHRTAWGNPLWNALHNLNIIGLCLAGSIITALIWPLALPVCLLFTLVTSVIFTLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETSPARGIFYVGYRRINDIGRELWLSMDDLTRHII
FFSTTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVRERKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFITQQSMILARDLGYRLE
GTDAQTLEVKKYKGRFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMMAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGLMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYIAIEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTEKTSRKRRKKHTEPYRIIDTFQDQLKRTQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
MSQHHVNPDLIHRTAWGNPLWNALHNLNIIGLCLAGSIITALIWPLALPVCLLFTLVTSVIFTLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETSPARGIFYVGYRRINDIGRELWLSMDDLTRHII
FFSTTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVRERKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFITQQSMILARDLGYRLE
GTDAQTLEVKKYKGRFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMMAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGLMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYIAIEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTEKTSRKRRKKHTEPYRIIDTFQDQLKRTQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 77096..123296
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| VJF88_RS00440 | 74815..77115 | - | 2301 | WP_029392471 | F-type conjugative transfer protein TrbC | - |
| VJF88_RS00445 | 77096..78214 | - | 1119 | WP_029391999 | DsbC family protein | trbB |
| VJF88_RS00450 | 78211..79554 | - | 1344 | WP_172686572 | conjugal transfer protein TrbA | trbA |
| VJF88_RS00455 | 79865..79948 | + | 84 | WP_271298668 | DinQ-like type I toxin DqlB | - |
| VJF88_RS00460 | 80011..80187 | - | 177 | WP_032172741 | hypothetical protein | - |
| VJF88_RS00465 | 80338..80805 | - | 468 | WP_000598962 | thermonuclease family protein | - |
| VJF88_RS00470 | 80965..81588 | + | 624 | WP_001155252 | helix-turn-helix domain-containing protein | - |
| VJF88_RS00475 | 81786..82001 | - | 216 | WP_000266543 | hypothetical protein | - |
| VJF88_RS00480 | 82005..82373 | - | 369 | WP_029391948 | hypothetical protein | - |
| VJF88_RS00485 | 82672..82914 | - | 243 | WP_029391947 | hypothetical protein | - |
| VJF88_RS00490 | 83243..83395 | + | 153 | WP_021532728 | Hok/Gef family protein | - |
| VJF88_RS00495 | 83709..83840 | - | 132 | Protein_98 | ethanolamine utilization protein EutE | - |
| VJF88_RS00500 | 83935..84144 | + | 210 | WP_000062946 | HEAT repeat domain-containing protein | - |
| VJF88_RS00505 | 84209..84862 | - | 654 | WP_029391946 | plasmid IncI1-type surface exclusion protein ExcA | - |
| VJF88_RS00510 | 84950..87115 | - | 2166 | WP_029391945 | DotA/TraY family protein | traY |
| VJF88_RS00515 | 87188..87757 | - | 570 | WP_000650755 | conjugal transfer protein TraX | - |
| VJF88_RS00520 | 87754..88959 | - | 1206 | WP_029391944 | conjugal transfer protein TraW | traW |
| VJF88_RS00525 | 88920..89537 | - | 618 | WP_029391943 | hypothetical protein | traV |
| VJF88_RS00530 | 89537..92581 | - | 3045 | WP_029391942 | conjugal transfer protein | traU |
| VJF88_RS00535 | 92757..93137 | - | 381 | WP_000690886 | hypothetical protein | - |
| VJF88_RS00540 | 93137..93445 | - | 309 | WP_029391941 | hypothetical protein | - |
| VJF88_RS00545 | 93641..93958 | + | 318 | WP_000118520 | quaternary ammonium compound efflux SMR transporter SugE | - |
| VJF88_RS00550 | 93955..94488 | - | 534 | WP_001221666 | lipocalin family protein | - |
| VJF88_RS00555 | 94582..95727 | - | 1146 | WP_000976514 | extended-spectrum class C beta-lactamase CMY-2 | - |
| VJF88_RS00560 | 96051..97313 | - | 1263 | WP_000608644 | IS1380-like element ISEcp1 family transposase | - |
| VJF88_RS00565 | 97650..98468 | - | 819 | WP_072132149 | conjugal transfer protein TraT | traT |
| VJF88_RS00570 | 98383..98634 | - | 252 | WP_021532719 | hypothetical protein | - |
| VJF88_RS00575 | 98691..99089 | - | 399 | WP_029391936 | DUF6750 family protein | traR |
| VJF88_RS00580 | 99136..99666 | - | 531 | WP_000987022 | conjugal transfer protein TraQ | traQ |
| VJF88_RS00585 | 99663..100376 | - | 714 | WP_021532717 | conjugal transfer protein TraP | traP |
| VJF88_RS00590 | 100373..101704 | - | 1332 | WP_323873455 | conjugal transfer protein TraO | traO |
| VJF88_RS00595 | 101708..102682 | - | 975 | WP_029391933 | DotH/IcmK family type IV secretion protein | traN |
| VJF88_RS00600 | 102693..103388 | - | 696 | WP_029391932 | DotI/IcmL family type IV secretion protein | traM |
| VJF88_RS00605 | 103400..103750 | - | 351 | WP_021532713 | hypothetical protein | traL |
| VJF88_RS00610 | 103767..107828 | - | 4062 | WP_029391931 | LPD7 domain-containing protein | - |
| VJF88_RS00615 | 107892..108182 | - | 291 | WP_000817799 | hypothetical protein | traK |
| VJF88_RS00620 | 108179..109327 | - | 1149 | WP_000625247 | plasmid transfer ATPase TraJ | virB11 |
| VJF88_RS00625 | 109311..110147 | - | 837 | WP_029391930 | type IV secretory system conjugative DNA transfer family protein | traI |
| VJF88_RS00630 | 110144..110596 | - | 453 | WP_001081199 | DotD/TraH family lipoprotein | - |
| VJF88_RS00635 | 110955..112157 | - | 1203 | WP_029391929 | conjugal transfer protein TraF | - |
| VJF88_RS00640 | 112247..113068 | - | 822 | WP_000804089 | hypothetical protein | traE |
| VJF88_RS00645 | 113319..113864 | - | 546 | WP_029391928 | hypothetical protein | - |
| VJF88_RS00650 | 113983..114264 | - | 282 | WP_029391927 | hypothetical protein | - |
| VJF88_RS00655 | 114305..115474 | - | 1170 | WP_029391926 | site-specific integrase | - |
| VJF88_RS00660 | 115529..115888 | + | 360 | WP_013307881 | hypothetical protein | - |
| VJF88_RS00665 | 115893..116282 | - | 390 | Protein_132 | phage tail protein | - |
| VJF88_RS00670 | 117234..117656 | + | 423 | WP_252347182 | hypothetical protein | - |
| VJF88_RS00675 | 117653..118900 | - | 1248 | WP_104209198 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| VJF88_RS00680 | 118897..119604 | - | 708 | WP_202110611 | A24 family peptidase | - |
| VJF88_RS00685 | 119564..120040 | - | 477 | WP_029391914 | lytic transglycosylase domain-containing protein | virB1 |
| VJF88_RS00690 | 120099..120635 | - | 537 | WP_029391915 | type 4 pilus major pilin | - |
| VJF88_RS00695 | 120686..121786 | - | 1101 | WP_029391916 | type II secretion system F family protein | - |
| VJF88_RS00700 | 121788..123296 | - | 1509 | WP_021532701 | GspE/PulE family protein | virB11 |
| VJF88_RS00705 | 123393..123989 | - | 597 | WP_029391917 | hypothetical protein | - |
| VJF88_RS00710 | 124085..124639 | - | 555 | WP_021532699 | hypothetical protein | - |
| VJF88_RS00715 | 124688..125140 | - | 453 | WP_029391918 | type IV pilus biogenesis protein PilP | - |
| VJF88_RS00720 | 125130..126425 | - | 1296 | WP_021532697 | type 4b pilus protein PilO2 | - |
| VJF88_RS00725 | 126446..128065 | - | 1620 | WP_021532696 | PilN family type IVB pilus formation outer membrane protein | - |
Host bacterium
| ID | 20052 | GenBank | NZ_CP141235 |
| Plasmid name | SB18002|unnamed | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 133297 bp | Coordinate of oriT [Strand] | 71437..71509 [-] |
| Host baterium | Escherichia sp. SB18002 |
Cargo genes
| Drug resistance gene | tet(A), dfrA1, blaTEM-1B, sul2, blaCMY-2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |