Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119609 |
| Name | oriT_pNCDO700A |
| Organism | Lactococcus cremoris strain NCDO700 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP069180 (3166..3203 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 1..7, 19..25 (ACACCAC..GTGGTGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pNCDO700A
ACACCACCCAATTTTGGAGTGGTGTGTAAGTGCGCATT
ACACCACCCAATTTTGGAGTGGTGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file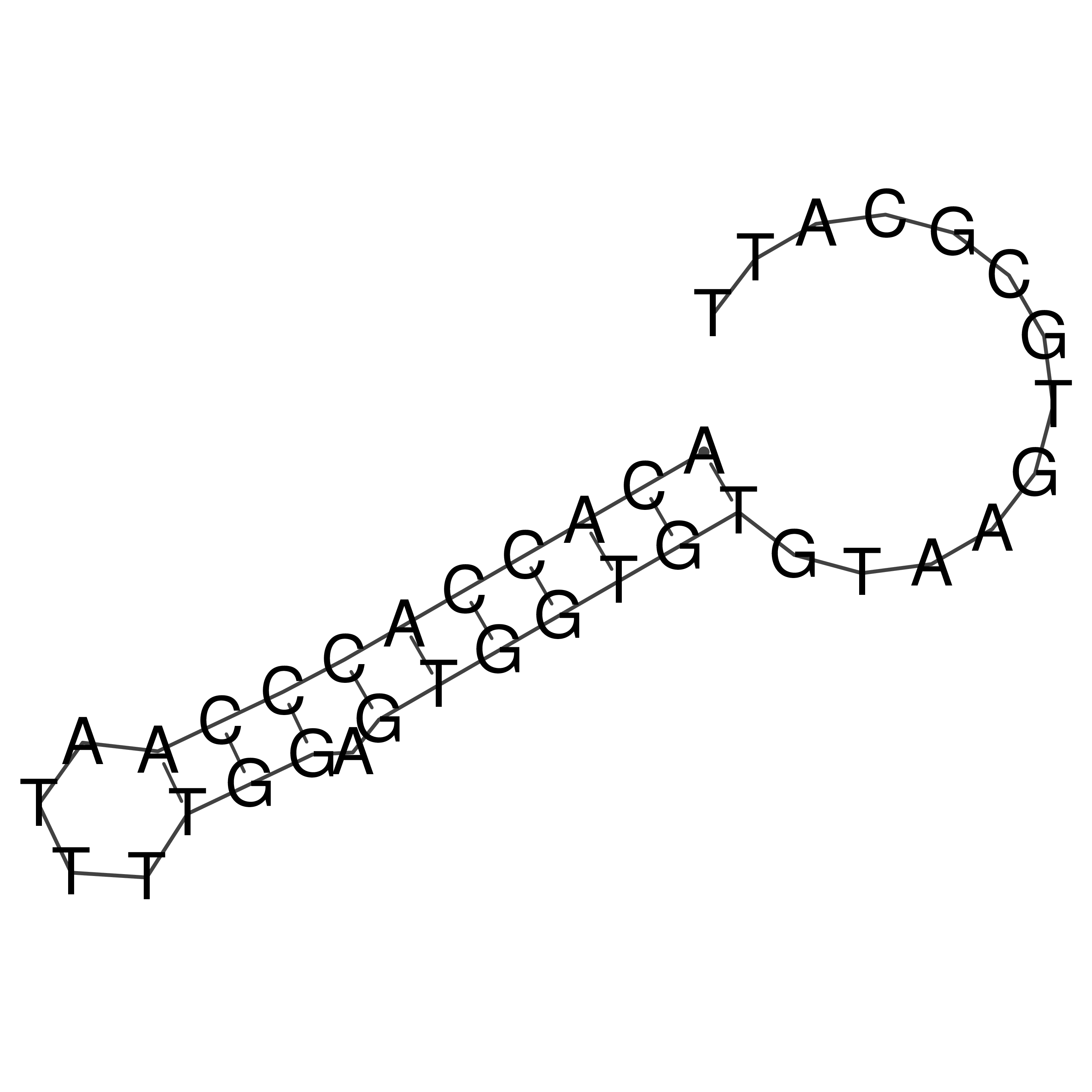
Relaxase
| ID | 12388 | GenBank | WP_010890626 |
| Name | mobQ_LLNCDO700_RS12980_pNCDO700A |
UniProt ID | A0A8B3ETK9 |
| Length | 680 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 680 a.a. Molecular weight: 79275.45 Da Isoelectric Point: 6.7052
>WP_010890626.1 MULTISPECIES: MobQ family relaxase [Lactococcus]
MAIFHMNFSNISAGKGRSAVASASYRSGEKLYSEMENKTYFYNRSVMPESFILLPENAPEWAKDRQKLWN
EVEAVDRKVNSRYAKEFNVALPIELSEDEQKELLTEYVQKIFVDKGMVADVAIHRDHDENPHAHVMLTNR
PFNADGSWGQKAKKEYILDENGNKTYTANGHARSRKIWLVDWDKVGKVEEWRKAWADHVNSVFQEKNIDE
RISEKTLEAQGINDIATQHVGVTGNRDERAEFNKLVLENRQHKAELENLDEKINNELKVKQLKNFYSFNE
KKVIAELSKELHTFIDLEHLEEKNKMLFNWKNSVLIKQIVGKDVSEELNKVSRQEFSLDNANRLIDKVVD
RSIKAMYPTVDSNAFSMPEKRQLIRETESENKVFTGAELDDRLSMIRADSLNRQIVALTKRPFTSLTMLS
KQEKYHIDNINNVLAYQGYNYENIEMSHGKILRNYESEEFSKELDIIQRSLKQINSIEEVRKIVEQQYSV
VLGTIFPDSELNKLSLVEKEKTYNAVIYFNPTIKKLTQSELENIVKEPPILFSTKEHNQGLSYLAGRIRL
EDINNKHLTRVLKFEGTKKLFIGEAKADKNIDPELLKEATNRNDKQSYKNEFYRDKNMENYQSVAYSDTS
PAEYMNRLFSGVLISVLYNNENQHKKGLEEVEWDMERKHREHTKTGGLSR
MAIFHMNFSNISAGKGRSAVASASYRSGEKLYSEMENKTYFYNRSVMPESFILLPENAPEWAKDRQKLWN
EVEAVDRKVNSRYAKEFNVALPIELSEDEQKELLTEYVQKIFVDKGMVADVAIHRDHDENPHAHVMLTNR
PFNADGSWGQKAKKEYILDENGNKTYTANGHARSRKIWLVDWDKVGKVEEWRKAWADHVNSVFQEKNIDE
RISEKTLEAQGINDIATQHVGVTGNRDERAEFNKLVLENRQHKAELENLDEKINNELKVKQLKNFYSFNE
KKVIAELSKELHTFIDLEHLEEKNKMLFNWKNSVLIKQIVGKDVSEELNKVSRQEFSLDNANRLIDKVVD
RSIKAMYPTVDSNAFSMPEKRQLIRETESENKVFTGAELDDRLSMIRADSLNRQIVALTKRPFTSLTMLS
KQEKYHIDNINNVLAYQGYNYENIEMSHGKILRNYESEEFSKELDIIQRSLKQINSIEEVRKIVEQQYSV
VLGTIFPDSELNKLSLVEKEKTYNAVIYFNPTIKKLTQSELENIVKEPPILFSTKEHNQGLSYLAGRIRL
EDINNKHLTRVLKFEGTKKLFIGEAKADKNIDPELLKEATNRNDKQSYKNEFYRDKNMENYQSVAYSDTS
PAEYMNRLFSGVLISVLYNNENQHKKGLEEVEWDMERKHREHTKTGGLSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A8B3ETK9 |
T4CP
| ID | 14547 | GenBank | WP_010890635 |
| Name | t4cp2_LLNCDO700_RS13185_pNCDO700A |
UniProt ID | _ |
| Length | 530 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 530 a.a. Molecular weight: 59615.30 Da Isoelectric Point: 4.8001
>WP_010890635.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Lactococcus]
MNGTILGVLDNKIIYQDNTTKPNRNVMVIGGSGSYKTQSVVITNLFNETKNSIVVTDPKGELYEKTAGIK
LAQGYEVHVVNFANMAHSDRYNPFDYIERDIQAESVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMK
ERSPEQRNLAGVINVLQTFDSEPINKDENSDLDNLFLALKITHPARIAYELGFKKAKGDMKASIISSLLA
TISKFTDEEVSNFTSISDFHLQDIGRKKIVLYVIIPVMDNTYESFINLFFSQMFDELYKLASSNGAKLPQ
EVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQSLYGKEKAESILGNHAVKICLNASNEA
TAKYFSELLGKSTVKVETGSESTSHSKETSTSKSDSYSYTSRQLMTPDEIIRMPDTQSLLIFTNQKPIKA
TKAFQFKLFPDADSKVKLEQNKYVGITSKSQLEKYNDLSVKWEEKLQSLKNITVTEEEEKDLQDNIDSDL
EAMNNENSGVDNSTTETDKIAETEAKEVQAEEKNFDQVVL
MNGTILGVLDNKIIYQDNTTKPNRNVMVIGGSGSYKTQSVVITNLFNETKNSIVVTDPKGELYEKTAGIK
LAQGYEVHVVNFANMAHSDRYNPFDYIERDIQAESVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMK
ERSPEQRNLAGVINVLQTFDSEPINKDENSDLDNLFLALKITHPARIAYELGFKKAKGDMKASIISSLLA
TISKFTDEEVSNFTSISDFHLQDIGRKKIVLYVIIPVMDNTYESFINLFFSQMFDELYKLASSNGAKLPQ
EVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQSLYGKEKAESILGNHAVKICLNASNEA
TAKYFSELLGKSTVKVETGSESTSHSKETSTSKSDSYSYTSRQLMTPDEIIRMPDTQSLLIFTNQKPIKA
TKAFQFKLFPDADSKVKLEQNKYVGITSKSQLEKYNDLSVKWEEKLQSLKNITVTEEEEKDLQDNIDSDL
EAMNNENSGVDNSTTETDKIAETEAKEVQAEEKNFDQVVL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 20039 | GenBank | NZ_CP069180 |
| Plasmid name | pNCDO700A | Incompatibility group | - |
| Plasmid size | 71195 bp | Coordinate of oriT [Strand] | 3166..3203 [-] |
| Host baterium | Lactococcus cremoris strain NCDO700 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |