Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119532 |
| Name | oriT_HAMBI 105|unnamed |
| Organism | Agrobacterium tumefaciens strain HAMBI 105 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP139999 (54844..54872 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_HAMBI 105|unnamed
AGGGCGCAATATACGTCGCTCGCGCGACG
AGGGCGCAATATACGTCGCTCGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file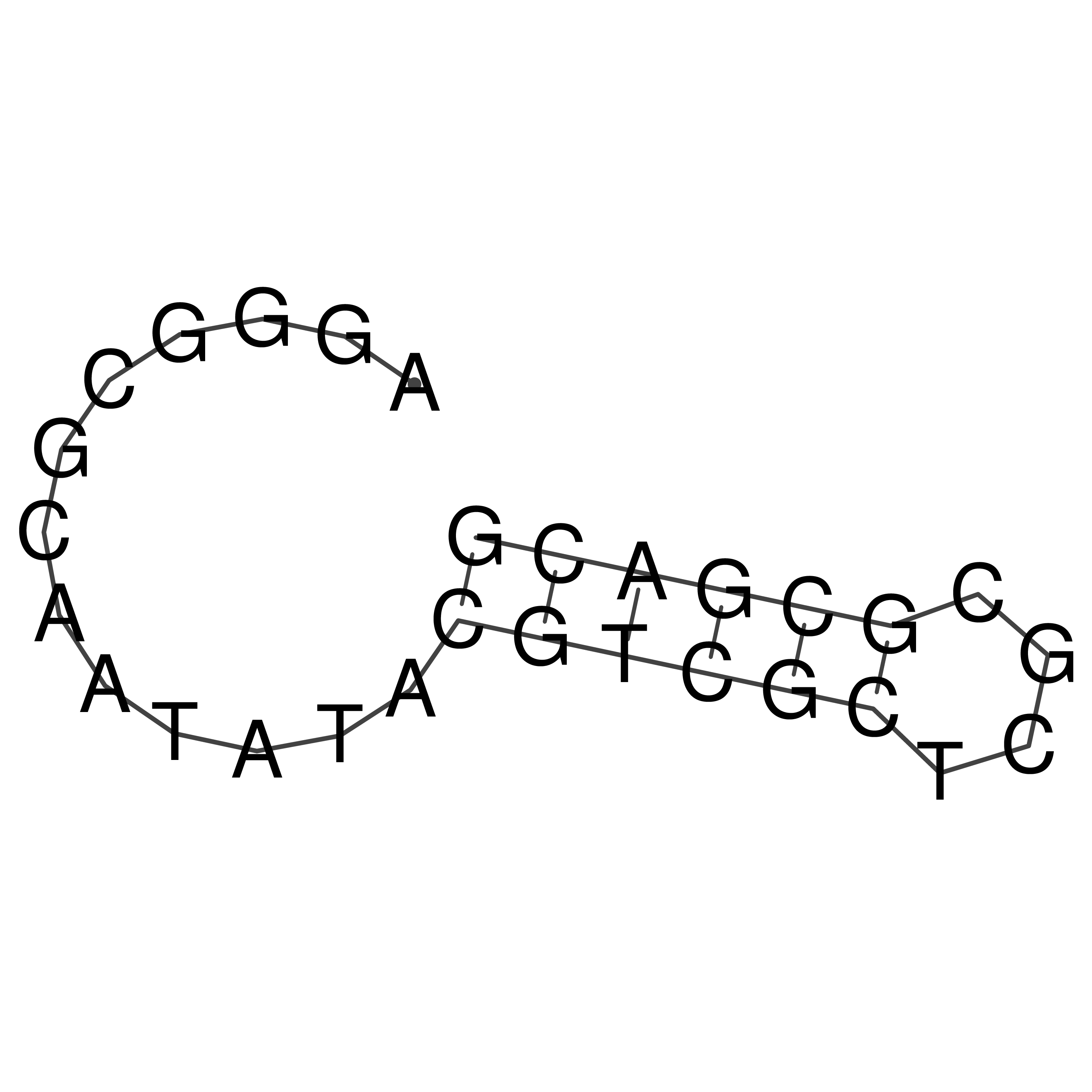
Relaxase
| ID | 12342 | GenBank | WP_114836199 |
| Name | traA_U0027_RS24110_HAMBI 105|unnamed |
UniProt ID | _ |
| Length | 1541 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1541 a.a. Molecular weight: 170827.34 Da Isoelectric Point: 7.8700
>WP_114836199.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium tumefaciens]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRARMMDEQVGTSFSYRGGASELVHEELALPDQIPAWLKSAIDG
KSVAGASEALWNAVDVFEKRADAQLARELIIALPEELTRAENIALVREFVADNFTSKGMVADWVFHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETMKTWKIAWADTANR
HLALACHEIRLDGRSYAEQGLAGIAQKHLGPEKAALARKGIERYFAPGDLARRQEIADRLLANPELLLKQ
LANERSTFDERDIARALHRYVDDPTDFANIRARLMASDDLVLLKPQQIDAETDKAAEPAVFTTREILRIE
YDMARSANVLSKRPGFAVSADAVAAAIRKVETEDPQKPFRLGPQQVDAVRHVTGDNGIAAVVGLAGAGKS
TLLAAARVAWESEGRRVIGAALAGKAAEGLEDSSGIRSRTLASLELAWASGRDLLERGNVFVIDEAGMIS
SQQMGRVLKVAEDAGAKVVLVGDAMQLQPIQAGAAFRAISDRIGFAELAGVRRQRQEWARDASRLFARGN
VRDGLDAYAQQGRITEAETRAEIVERIVADWTDARRNLLQNTTDRGDAPRLRGDELLVLAHTNEDVKRLN
ESLRKVMMEEGVLTDARAFQTARGAREFAAGDRIIFLENARFVEPRARHLGPQYVKNGMLGTVVSTGDKR
GGSLLSIRLDNSRDVVISEDSYRNIDHGYAATIHKSQGATIDRTFVLATGMMDQHLTYVSMTRHRIRADL
YAAREDFELRPEWGRKPRADHAAGVTGELVETGEAKFRPQDEDAKQSPYADVKTDNGAVHRLWGVSLPKA
LEEGGISEGDTVTLRKDGVERVKVKVTVVDEQTGEKRAEEREVERNIWTAELKETADAREQRIEQESHRP
ELFSQLVDRLGRSGAKTTTLDFESEEGYRAHAEDFARRRGIDMAAGIGEGVSGRLAWIAEKRGQVAKLWE
RASIALGFAIERERRIAYNEDRTETPAPEIPTTGQYLIPPTTRFARGVDEDARRAQLASERWKERDAILG
PVLEKIYRDPDAALARLNALASDATNEPRKFADDLAAAPDRLGRLRGSDLMVDGRAARDERKTATAALSE
LVPMARAHATEFRRQAERFGIREEQRRAHMSLSIPMLSKPAMARLVEIEEIRERGGDDAYKTAFAYAVED
RQLVQEVKAVTDALTARFGWSAFTTKADAVAERNMAERMPEDLAPERREKLLRLFTVVRRFSEEQHLAEK
QDRSKIVAGASVEVGQEAIPMLPMLAAVTEFKTPVEDEARIRAGAVAHYRHHRAALADTAALIWRDPAGA
VAKIEELVGKGFAGERIAAAVSNDPAAYGALRGSDRMMDRMFAAGRERKEAVQAVPEAASRVRSLGASWA
IALDTETRAVTEERRRMAVAVPGLSQTAEDALRTLVADVRKQKKDKGENIRRDVAARSLDPRIIREFAAV
SRALDERFGRNAILRGEKDVVNRVSPAQRRAFEAMRERLLILQQVVRTTSSQEIIAERQRQDIDRTRGFT
R
MAIMFVRAQVIGRGAGRSIVSAAAYRHRARMMDEQVGTSFSYRGGASELVHEELALPDQIPAWLKSAIDG
KSVAGASEALWNAVDVFEKRADAQLARELIIALPEELTRAENIALVREFVADNFTSKGMVADWVFHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETMKTWKIAWADTANR
HLALACHEIRLDGRSYAEQGLAGIAQKHLGPEKAALARKGIERYFAPGDLARRQEIADRLLANPELLLKQ
LANERSTFDERDIARALHRYVDDPTDFANIRARLMASDDLVLLKPQQIDAETDKAAEPAVFTTREILRIE
YDMARSANVLSKRPGFAVSADAVAAAIRKVETEDPQKPFRLGPQQVDAVRHVTGDNGIAAVVGLAGAGKS
TLLAAARVAWESEGRRVIGAALAGKAAEGLEDSSGIRSRTLASLELAWASGRDLLERGNVFVIDEAGMIS
SQQMGRVLKVAEDAGAKVVLVGDAMQLQPIQAGAAFRAISDRIGFAELAGVRRQRQEWARDASRLFARGN
VRDGLDAYAQQGRITEAETRAEIVERIVADWTDARRNLLQNTTDRGDAPRLRGDELLVLAHTNEDVKRLN
ESLRKVMMEEGVLTDARAFQTARGAREFAAGDRIIFLENARFVEPRARHLGPQYVKNGMLGTVVSTGDKR
GGSLLSIRLDNSRDVVISEDSYRNIDHGYAATIHKSQGATIDRTFVLATGMMDQHLTYVSMTRHRIRADL
YAAREDFELRPEWGRKPRADHAAGVTGELVETGEAKFRPQDEDAKQSPYADVKTDNGAVHRLWGVSLPKA
LEEGGISEGDTVTLRKDGVERVKVKVTVVDEQTGEKRAEEREVERNIWTAELKETADAREQRIEQESHRP
ELFSQLVDRLGRSGAKTTTLDFESEEGYRAHAEDFARRRGIDMAAGIGEGVSGRLAWIAEKRGQVAKLWE
RASIALGFAIERERRIAYNEDRTETPAPEIPTTGQYLIPPTTRFARGVDEDARRAQLASERWKERDAILG
PVLEKIYRDPDAALARLNALASDATNEPRKFADDLAAAPDRLGRLRGSDLMVDGRAARDERKTATAALSE
LVPMARAHATEFRRQAERFGIREEQRRAHMSLSIPMLSKPAMARLVEIEEIRERGGDDAYKTAFAYAVED
RQLVQEVKAVTDALTARFGWSAFTTKADAVAERNMAERMPEDLAPERREKLLRLFTVVRRFSEEQHLAEK
QDRSKIVAGASVEVGQEAIPMLPMLAAVTEFKTPVEDEARIRAGAVAHYRHHRAALADTAALIWRDPAGA
VAKIEELVGKGFAGERIAAAVSNDPAAYGALRGSDRMMDRMFAAGRERKEAVQAVPEAASRVRSLGASWA
IALDTETRAVTEERRRMAVAVPGLSQTAEDALRTLVADVRKQKKDKGENIRRDVAARSLDPRIIREFAAV
SRALDERFGRNAILRGEKDVVNRVSPAQRRAFEAMRERLLILQQVVRTTSSQEIIAERQRQDIDRTRGFT
R
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14485 | GenBank | WP_114836202 |
| Name | traG_U0027_RS24095_HAMBI 105|unnamed |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70501.47 Da Isoelectric Point: 9.4541
>WP_114836202.1 Ti-type conjugative transfer system protein TraG [Agrobacterium tumefaciens]
MALKGKLHPSLLLVFVPIVVTAISVYIVGWRWKGLATGMSGKTAYWFLRATPLPALLFGPLAGLLTVSAL
PMHRRRPIAMVSLLCFLGVVGFYALREYGRLAPSVGRGVVSWDYALSYLDMVAVVGALCGFMAVAASARI
STVVPEQVKRARRATFGDADWLSMSAAGKLFPADGEIVVGERYRVDKEIVHELPFDPNDRSTWGQGGKAP
LLTYNQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGVAHMLLSESMRFESSTGSYFQSQAHNLLTGLLDHVMLSPDYAGRRN
LRSLRQIVSEPEPSVLTMLRDIQENSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGRKDVFLNIPASILRSYPGIGRVIIGALINAMTQADGAFKRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDMKNSASRKSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDAAAR
VNRFVKLGS
MALKGKLHPSLLLVFVPIVVTAISVYIVGWRWKGLATGMSGKTAYWFLRATPLPALLFGPLAGLLTVSAL
PMHRRRPIAMVSLLCFLGVVGFYALREYGRLAPSVGRGVVSWDYALSYLDMVAVVGALCGFMAVAASARI
STVVPEQVKRARRATFGDADWLSMSAAGKLFPADGEIVVGERYRVDKEIVHELPFDPNDRSTWGQGGKAP
LLTYNQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGVAHMLLSESMRFESSTGSYFQSQAHNLLTGLLDHVMLSPDYAGRRN
LRSLRQIVSEPEPSVLTMLRDIQENSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGRKDVFLNIPASILRSYPGIGRVIIGALINAMTQADGAFKRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDMKNSASRKSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDAAAR
VNRFVKLGS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 16032..25643
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| U0027_RS23835 (U0027_23830) | 12088..12375 | + | 288 | WP_080821105 | hypothetical protein | - |
| U0027_RS23840 (U0027_23835) | 12382..12768 | + | 387 | WP_114836239 | hypothetical protein | - |
| U0027_RS23845 (U0027_23840) | 12806..13519 | - | 714 | WP_114836238 | IS6 family transposase | - |
| U0027_RS23850 (U0027_23845) | 13916..14113 | - | 198 | WP_162791770 | hypothetical protein | - |
| U0027_RS23855 (U0027_23850) | 14131..14574 | - | 444 | WP_114836235 | hypothetical protein | - |
| U0027_RS23860 (U0027_23855) | 15041..15391 | - | 351 | WP_114836234 | MarR family transcriptional regulator | - |
| U0027_RS23865 (U0027_23860) | 15490..16029 | + | 540 | WP_114836233 | hypothetical protein | - |
| U0027_RS23870 (U0027_23865) | 16032..16706 | + | 675 | WP_114836232 | lytic transglycosylase domain-containing protein | virB1 |
| U0027_RS23875 (U0027_23870) | 16703..17002 | + | 300 | WP_114836231 | TrbC/VirB2 family protein | virB2 |
| U0027_RS23880 (U0027_23875) | 17009..17347 | + | 339 | WP_114836230 | type IV secretion system protein VirB3 | virB3 |
| U0027_RS23885 (U0027_23880) | 17340..19706 | + | 2367 | WP_114836229 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| U0027_RS23890 (U0027_23885) | 19703..20404 | + | 702 | WP_114836228 | P-type DNA transfer protein VirB5 | virB5 |
| U0027_RS23895 (U0027_23890) | 20401..20634 | + | 234 | WP_114836227 | EexN family lipoprotein | - |
| U0027_RS23900 (U0027_23895) | 20639..21571 | + | 933 | WP_114836226 | type IV secretion system protein | virB6 |
| U0027_RS23905 (U0027_23900) | 21611..21892 | + | 282 | WP_114836225 | hypothetical protein | - |
| U0027_RS23910 (U0027_23905) | 21894..22565 | + | 672 | WP_114836224 | virB8 family protein | virB8 |
| U0027_RS23915 (U0027_23910) | 22562..23419 | + | 858 | WP_114836223 | P-type conjugative transfer protein VirB9 | virB9 |
| U0027_RS23920 (U0027_23915) | 23428..24603 | + | 1176 | WP_114836222 | type IV secretion system protein VirB10 | virB10 |
| U0027_RS23925 (U0027_23920) | 24612..25643 | + | 1032 | WP_114836221 | P-type DNA transfer ATPase VirB11 | virB11 |
| U0027_RS23930 (U0027_23925) | 25852..26550 | + | 699 | WP_236762449 | hypothetical protein | - |
| U0027_RS23935 (U0027_23930) | 26547..27134 | + | 588 | WP_114836220 | RES family NAD+ phosphorylase | - |
| U0027_RS23940 (U0027_23935) | 27367..27573 | + | 207 | WP_162791769 | hypothetical protein | - |
| U0027_RS23945 (U0027_23940) | 27670..27930 | + | 261 | WP_114836219 | hypothetical protein | - |
| U0027_RS23950 (U0027_23945) | 28114..28298 | - | 185 | Protein_33 | transposon DNA-invertase | - |
Host bacterium
| ID | 19962 | GenBank | NZ_CP139999 |
| Plasmid name | HAMBI 105|unnamed | Incompatibility group | - |
| Plasmid size | 410809 bp | Coordinate of oriT [Strand] | 54844..54872 [+] |
| Host baterium | Agrobacterium tumefaciens strain HAMBI 105 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |